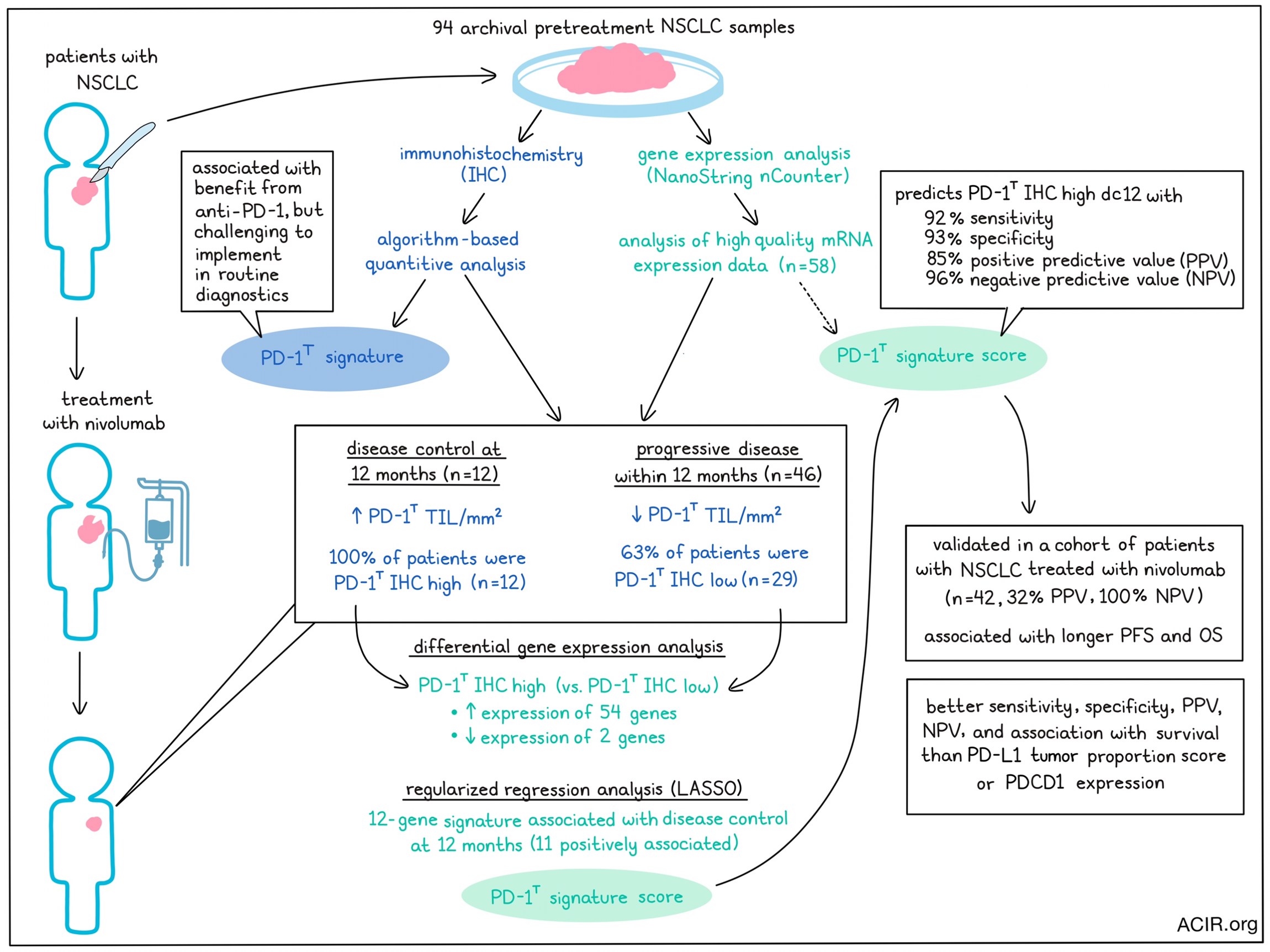
Despite its successes in some patients, immune checkpoint blockade (ICB) targeting PD-1 is not beneficial in most patients with non-small cell lung cancer (NSCLC). Determining which patients would or would not benefit from this treatment has remained a challenge. Previously, a dysfunctional T cell subset has been associated with response to treatment, but this subset is not easily detectable in the clinical setting. Therefore, Hummelink et al. developed a gene signature to serve as a diagnostic tool in a recent publication in Clinical Cancer Research.
The researchers previously reported that a subset of tumor-infiltrating PD-1+ T cells with a dysfunctional phenotype, named PD-1T, were reactive to tumor cells and associated with long-term benefit from PD-1 blockade in NSCLC. These cells can be analyzed using an algorithm-based quantitative analysis of immunohistochemistry (IHC) samples from formalin-fixed paraffin-embedded (FFPE) tissues. However, this method proved to be challenging to implement in routine diagnostics for clinical care.
To develop a predictive mRNA signature reflecting PD-1T tumor infiltration, 94 archival pretreatment samples were selected from patients with advanced-stage NSCLC treated with nivolumab (training cohort). Sufficient RNA could be extracted from 70% of samples for gene expression analysis using the NanoString nCounter platform. High-quality mRNA expression data of 62% of samples (n=58) could be used for the development of the gene signature.
The endpoint of disease control at 12 months (DC 12m) was used to determine the benefit of treatment. Of the 58 patients, 12 showed DC 12m, while 46 developed progressive disease (PD) within 12 months of treatment. Using IHC data, the researchers confirmed that the number of PD-1T TIL per mm2 was significantly higher in the patients with DC 12m than in those with PD, consistent with their prior results. All patients with DC 12m were classified as PD-1T IHC high, as compared to only 37% of patients with PD.
To develop the mRNA expression gene set with maximum contrast, the researchers performed differential gene expression analysis between PD-1T IHC high patients with DC 12m (n=12) and PD-1T IHC low patients with PD (n=29). This revealed 54 genes that were more highly expressed and 2 genes that were lower in expression in the PD-1T IHC high DC 12m group. Upregulated genes included LAG3, CTLA4, CXCR6, and CXCL13, and genes related to type I interferon signaling, lymphocyte chemotaxis regulation, and NK cell-mediated cytotoxicity pathways. Regularized regression analysis (LASSO) revealed a 12-gene signature (the PD-1T signature score), which included STAT1, OAS1, TAP1, HEY1, CXCL13, IFIT2, IL6, TDO2, CD6, CD274, and LAG3. This PD-1T signature score was the most predictive, with all genes, except for HEY1, being positively associated with DC 12m.
There was a high correlation between the number of PD-1T tumor-infiltrating lymphocytes assessed by IHC and the PD-1T signature score. Using the signature, the researchers could separate the preselected PD-1T IHC high DC 12m group from the PD group. The area under the ROC curve (AUC) was 0.97, and using a probability score of 0.35 as the optimal cutoff, the sensitivity was 92%, with a specificity of 93%, a positive predictive value (PPV) of 85%, and a negative predictive value (NPV) of 96%.
To validate the signature, an independent cohort of 42 patients with advanced-stage NSCLC treated with nivolumab was assessed, of whom 14% (n=6) had DC 12m. In this validation cohort, the IHC PD-1T signature scores were higher in the DC 12m group than in the PD group, as in the training cohort. Validating the signature score, a high PD-1T signature score correctly identified all 6 patients with DC 12m, and a low score identified 23 of 36 patients with PD, resulting in a PPV of 32% and an NPV of 100% with an AUC of 0.87. Progression-free survival (PFS) and overall survival (OS) were longer in patients with high PD-1T signature scores.
The PD-L1 tumor proportion score (TPS) has previously been used as a predictor of therapy outcome. Therefore, the researchers compared the PD-L1 TPS with the PD-1T signature in the validation cohort. PD-L1 TPS was not significantly higher in those with DC 12m compared to patients with PD, and the AUC was lower than for the PD-1T signature. At a cutoff of 50%, the sensitivity, specificity, PPV, and NPV were lower than those associated with the PD-1T signature. Further, the TPS was not associated with PFS, and only borderline significant with OS. When using 1% as a cutoff, sensitivity, NPV, and survival data were similar to the 50% cutoff, while specificity and PPV were slightly lower. Therefore, the PD-1T signature better predicted outcomes than PD-L1 TPS.
Another factor that might be predictive is the gene expression of PDCD1, which encodes PD-1. Comparing the expression of this gene with the PD-1T signature, the researchers found that in the 58 pretreatment samples, the expression of PDCD1 was slightly higher in the DC 12m group than in the PD group. The AUC was lower than the PD-1T signature, as were sensitivity, NPV, and specificity, and patients with high PDCD1 scores did not have prolonged PFS or OS.
Overall, these data show that the PD-1T signature derived in this study is more accurate in predicting DC 12m and survival than PD-L1 TPS or PDCD1 expression. This confirms previous data that a specific PD-1+ T cell subset is predictive for anti-PD-1 outcomes, and not the total PD-1+ population. Once this signature is validated in larger cohorts, it may aid in selecting patients who would not benefit from PD-1 blockade monotherapy, preventing a potentially toxic or wasteful treatment and allowing for consideration of other therapies or combination strategies.
Write-up by Maartje Wouters, image by Lauren Hitchings
Meet the researcher
This week, first author Karlijn Hummelink and lead author Kim Monkhorst answered our questions.

What was the most surprising finding of this study for you?
The therapeutic efficacy of PD-1 blockade is confined to a minority of patients with advanced stage NSCLC, emphasizing the need for biomarkers to guide treatment decisions. Our previous work established PD-1T TILs, a dysfunctional tumor-infiltrating lymphocyte (TIL) pool with tumor-reactive potential, as a novel predictive biomarker for long-term benefit to PD-1 blockade with a high negative predictive value (NPV). However, the assessment of PD-1T TILs through advanced digital image analysis-based quantification of IHC stainings is complex, limiting its use in routine patient care. In our current study, we are pleased to report the successful translation of the PD-1T TIL signal into a PD-1T mRNA signature. The signature was developed using the Nanostring nCounter platform, a well established technology recognized for its robustness. This approach enhances the applicability of the biomarker, making it more suitable for implementation into diagnostic procedures.
What is the outlook?
This retrospective study established a clinically applicable PD-1T mRNA signature, demonstrating comparable predictive performance to the digital IHC quantification method. The results underscore the broader implications of our approach, with regard to integrating expression-level biomarkers into diagnostic procedures to improve therapeutic decision-making. The next steps will involve validating this biomarker in larger prospective trials, particularly within the current clinical setting of dual treatment with PD-1 blockade and chemotherapy. Additionally, there is keen interest in evaluating the predictive performance of the PD-1T signature in other cancer types treated with PD-1 blockade.
What was the coolest thing you’ve learned (about) recently outside of work?
KH: I am very passionate about cooking. As a pathology resident and researcher, cooking serves as my therapeutic escape after a long day at work, allowing me to unwind and reset my mind. My culinary preferences are the Mediterranean and Asian cuisines. Recently, I also did a wine course, exploring the origins of wine and acquiring knowledge about diverse flavors to enhance my skills in food pairing.
KM: learning all the different Padel shots and their names, vamos!




