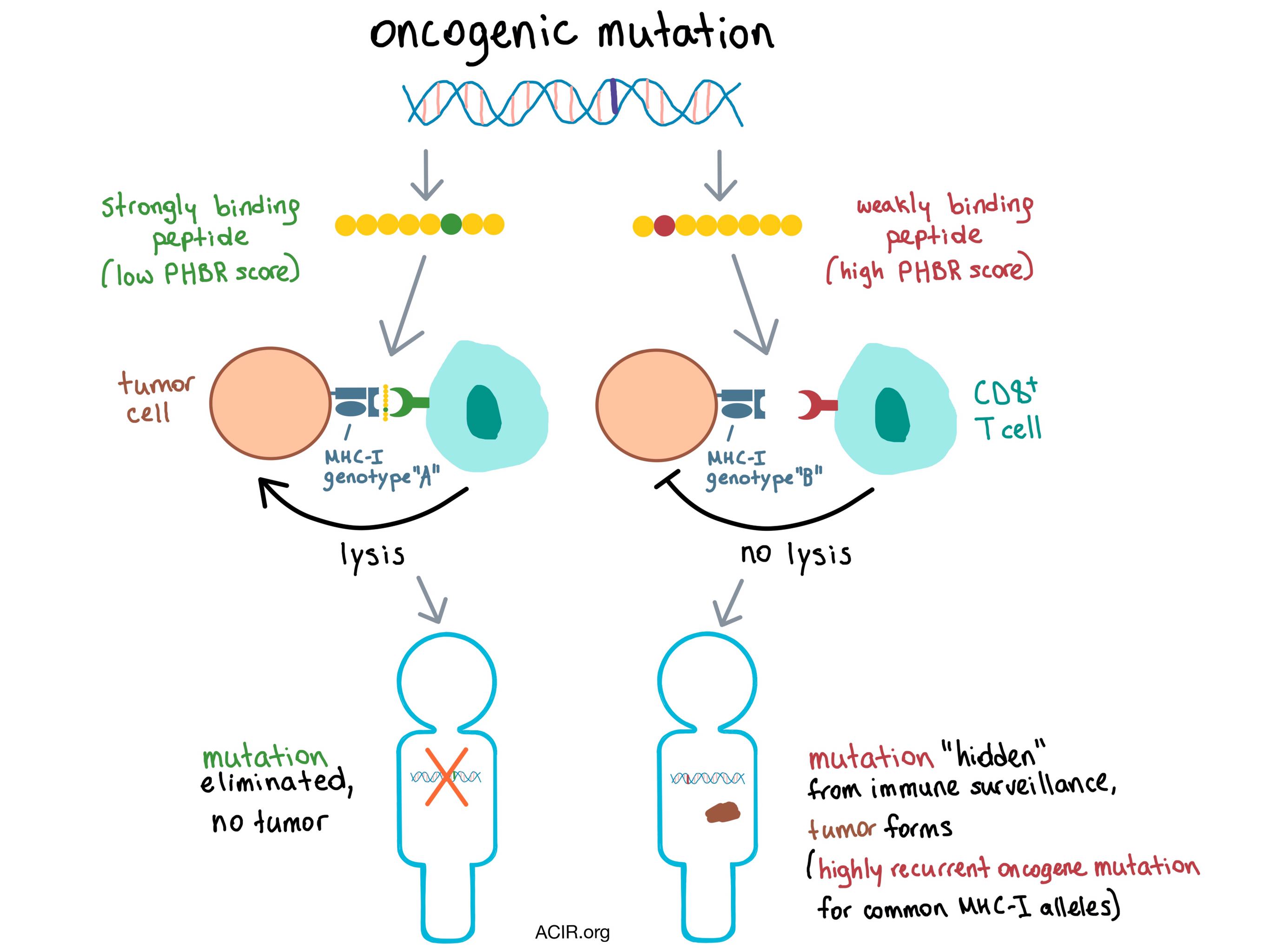
How does the immune system sculpt the tumor genome even before the tumor adapts counteracting immune evasion mechanisms? Marty et al. hypothesized that antigenic oncogenic mutations are eliminated by the immune system in the early stages of tumorigenesis, and this process is dependent on which oncogenic peptides can be effectively presented by an individual’s MHC-I molecules.
CD8+ T cells recognize target cells via target cell-derived epitope-length peptides bound to an MHC-I molecule on the cell surface. The binding affinity between the MHC-I complex and a peptide depends on the amino acid sequence of the peptide and the structure of the presenting MHC molecule, which is based on the three genes that encode the human leukocyte antigen (HLA, aka MHC): HLA-A, HLA-B, and HLA-C. Each individual has two alleles per gene, for a total of six alleles, and the HLA locus is highly polymorphic, with >10,000 alleles documented to date. Taking this heterogeneity into account, Marty et al. developed a qualitative, predicted binding affinity-based, residue-centric score that they used to classify whether a peptide sequence containing a specific amino acid residue would bind weakly or strongly to the MHC-I molecule. The researchers evaluated several strategies and found that the best predictor of presentation of a certain residue was the “best rank” score, indicating the lowest score (highest binding affinity) among all 8-11mer peptides containing that residue, and this was used in all subsequent analyses. This score was validated experimentally by comparing mutations in 5 cancer cell lines that were predicted to be presented by MHC-I with peptides that actually bound to MHC-I, as identified by mass spectrometry (MS).
To find out whether differences in MHC-I genotypes resulted in differential presentation of mutations in individual patients, Marty et al. analyzed a large human cancer cohort using sequencing data from The Cancer Genome Atlas (TCGA). Using several algorithms, the team assigned HLA-A, HLA-B, and HLA-C allele pairs to each patient, and predicted and ranked the binding affinities of all the somatic mutations within each tumor. A Patient Harmonic-mean Best Rank (PHBR) score was calculated, reflecting a patient’s ability to present a specific mutation. The PHBR score is dominated by the best ranking allele, but also takes into account the binding affinities of the other five alleles. The predictive power of the PHBR score was validated using independent MS data. The researchers used these tools to analyze a set of over 1,000 known oncogene and tumor suppressor driver mutations and found that some recurrent oncogenic mutations are poorly presented by MHC-I across the board.
The researchers hypothesized that since the MHC-I genotype determines which oncogenic peptides are presented to the immune system, mutations that are exposed to immune surveillance (i.e., mutations creating peptides that would bind to the patient-specific MHC-I) should be eliminated early on and observed less frequently than the mutations “hidden” from immune surveillance due to poor presentation by patient-specific MHC-I. Indeed, an analysis of the relationship between PHBR and mutation probability revealed that patients were more likely to acquire mutations that were less effectively presented by their MHC-I. Therefore, the PHBR score predicted which recurrent oncogenic mutations were more likely to drive early tumor development in individuals across many cancer types. Perhaps unsurprisingly, patients with HLA mutations, and therefore an impaired ability to present peptides via MHC-I, had a significantly higher overall number of mutations in the tumor, further supporting the hypothesis that MHC-I genotype restriction is a “gatekeeper” of immunoediting during early tumorigenesis.
To gain a big-picture perspective on how MHC-I genotype affects the frequencies of tumor mutations on a population level, the team took another look at the TCGA data and found that the population’s ability to present a certain mutation was significantly and inversely correlated with that mutation’s frequency in tumors. After further analysis, Marty et al. concluded that poor presentation of peptides containing highly recurrent oncogenic mutations is a general feature of the majority of human MHC-I alleles.
This research demonstrates that an individual’s MHC-I genotype contributes to immunoediting of the common oncogenes and tumor suppressors in the early stages of tumor development. The mutation-centric presentation scores developed by Marty et al. can be combined with other known risk factors to potentially predict an individual’s susceptibility to developing a certain type of cancer.
by Anna Scherer




