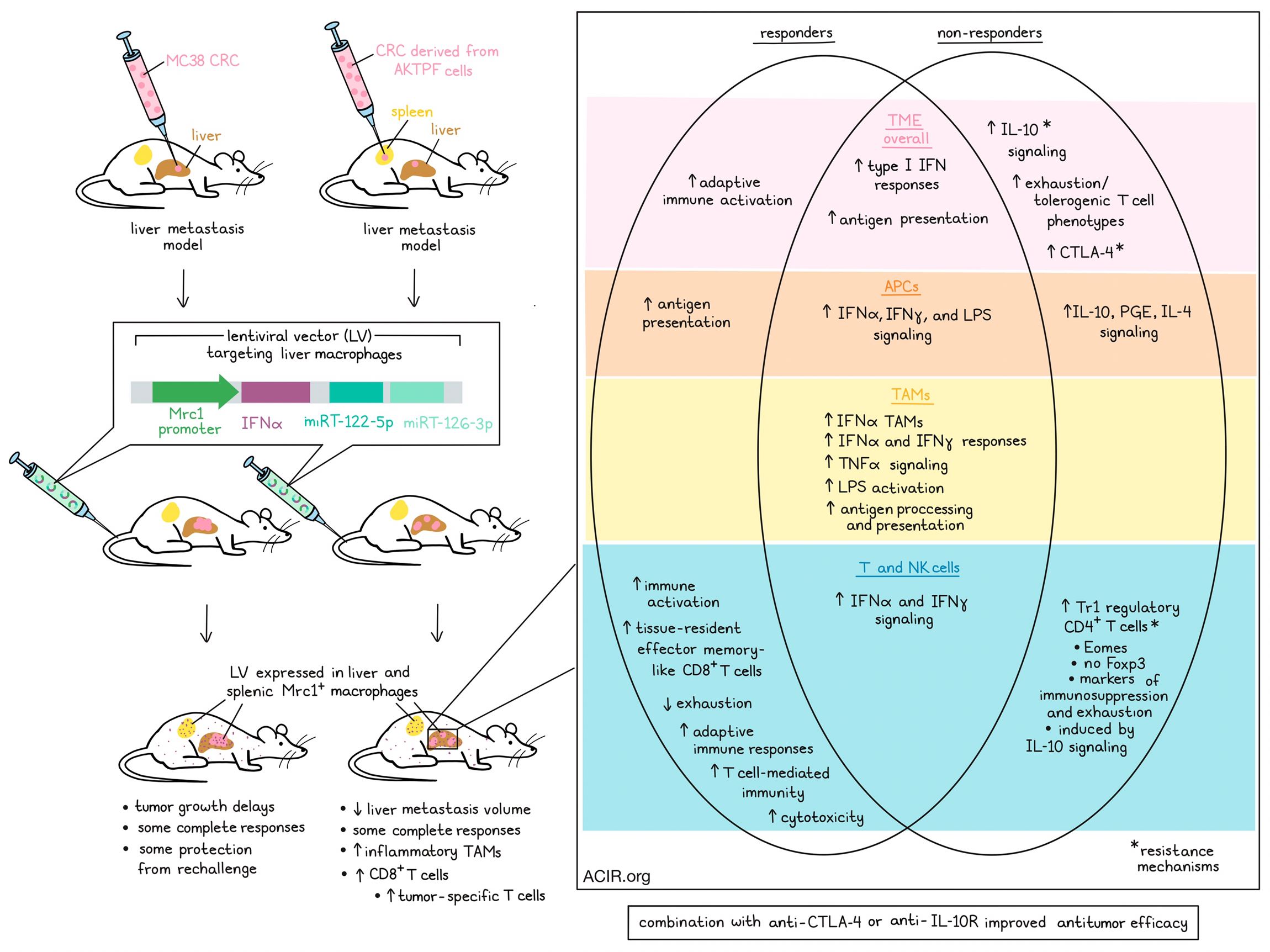
Liver metastases respond poorly to current immunotherapies, which may be due to the immunosuppressive tumor microenvironment (TME), and in particular, the presence of pro-tumoral tumor-associated macrophages (TAMs). To overcome this issue, Kerzel, Giacca, et al. hypothesized that enforced IFNα expression in liver metastases by genetic engineering of liver macrophages, including Kupffer cells (KCs) and TAMs, might reprogram the TME and enhance antitumor immune responses. Their assessment of this method in preclinical liver metastasis models was recently published in Cancer Cell.
The researchers first established whether they could specifically target liver metastasis macrophages with a lentiviral vector (LV) by generating LVs containing a putative promoter sequence from the mouse Mrc1 gene and a downstream GFP coding sequence. Immunocompromised mice were injected intravenously (i.v.) with these LVs, resulting in GFP expression in liver and spleen cells. To target the gene expression more specifically to macrophages, miRT-122-5p and miRT-126-3p were incorporated downstream of GFP to create Mrc1.GFP.miRT LV. Mice were injected intrahepatically with MC38 colorectal cancer (CRC) cells or intrasplenically with CRC cells derived from APCΔ716; KrasG12D; Tgfbr2-/-; Trp53R270H; Fbxw7-/- mice (AKTPF cells) to mimic metastatic seeding to the liver. Mice were then treated with Mrc1.GFP.miRT LV by i.v. injection. The presence of the miRNA regulation resulted in specific expression of the LV in liver macrophages and splenic MRC1+ macrophages.
These methods were then used to engineer macrophages to deliver IFNα to liver metastases by replacing the GFP with an IFNα-coding DNA sequence (IFNα LV). These IFNα LV were then injected i.v. in immunocompetent mice, resulting in a rapid transgene output, as observed by increasing IFNα concentrations in plasma. No hepatotoxicity or other treatment-related abnormalities were detected.
To assess the effects of local macrophage-delivered IFNα in the TME, mice previously challenged with MC38-based liver metastases were treated with IFNα LVs. Treatment delayed tumor progression, including a few complete responses (CR). When these CR mice were re-challenged with subcutaneous MC38 tumors, tumor growth was impaired, suggestive of induction of memory responses.
Assessing the effects of IFNα on the TME, AKTPF CRC liver metastases were treated with IFNα LV. This reduced the volume of liver metastases, induced CRs in 8/20 mice, increased in the proportion of TAMs with an inflammatory phenotype, and increased the number of CD8+ T cells.
To determine whether tumor-reactive T cells were induced in the TME in response to the macrophage engineering, syngeneic immunocompetent mice with established OVA-expressing MC38 liver metastases were treated with IFNα LV, and tumors were harvested early after treatment. OVA-specific T cells were enriched in tumors of treated mice, and the IFNα antitumor effects were reduced when CD8+ T cells were depleted. To determine whether the IFNα produced by engineered macrophages directly activated CD8+ T cells, the researchers used transgenic mice with IFNαR1 KO in CD4+ and CD8+ T cells. In these mice, the therapeutic effects of IFNα LV were maintained, suggesting that IFNα indirectly activated T cells.
To determine the mechanisms behind the antitumor effects, the researchers performed transcriptomic analyses on AKTPF liver metastases after treatment. Mice were assigned to three cohorts: controls, responders (CR were not included due to a lack of tissue), and resistant mice. Genes related to type I interferon responses were enriched in liver metastases and peri-metastatic areas in the IFNα LV-treated mice. In responders, genes related to adaptive immune activation were upregulated. Genes related to antigen presentation were highly expressed in responders and resistant mice. In the resistant mice, increased IL-10 signaling was observed, potentially responsible for the lack of therapeutic effects. Furthermore, markers of exhaustion and tolerogenic T cell phenotypes were also upregulated in the resistant mice.
Metastatic areas were subjected to single-cell transcriptomics and clustered into distinct cell types. Zooming in on the APC cluster, Kerzel, Giacca, et al. found that genes related to IFNα, IFNγ, or LPS signaling were enriched in treated groups. Genes related to IL-10, PGE2, and IL-4 signaling were only upregulated in the resistant group, while genes related to antigen presentation were upregulated in the responders. In both treatment groups, the TAM cluster was reshaped by IFNα treatment. These IFNα-TAMs had upregulated genes related to IFNα/IFNγ response, TNFα signaling, LPS activation, and antigen processing and presentation.
In the T and NK cell compartment, genes related to IFNα and IFNγ signaling were enriched in all treated groups, while genes related to immune activation were only enriched in responders. In resistant mice, there was an enriched population of regulatory CD4+ T cells (Tr1) that expressed Eomes and markers of exhaustion and immune suppression (including Ctla4), but lacked Foxp3. In responders, there was enrichment of a CD8+ T cell population resembling tissue-resident effector memory cells. Genes related to T cell exhaustion were downregulated in responders, while genes related to adaptive immune responses, T cell-mediated immunity, and cytotoxicity were upregulated.
To determine whether IFNα signaling in human CRC liver metastases was positively associated with the presence of Eomes+ CD4+ T cells in the TME, bulk RNAseq of human CRC liver metastases was performed. Patients with a higher IFNα signaling score had higher levels of the Tr1 signature score. Furthermore, a positive relationship between the IFNα score and Tr1 score, CTLA-4, and HLA-C expression was detected in metastatic areas. Genes involved in antigen presentation and immune cell activation were also positively correlated with the IFNα score. Immunostaining of CRC liver metastases samples showed that those with high IFNα had higher proportions of LAG3+CD4+ T, LAG3+EOMES+CD4+ T cells, and CTLA-4+CD4+ T cells in metastases.
When IL-10 signaling was inhibited with an IL-10R blocking antibody in mice with AKTPF metastases treated with IFNα LV, it blocked the accumulation of Eomes+ CD4+ T cells, and improved therapeutic effects. Since Ctla4 was upregulated in resistant mice, combined IFNα LV and anti-CTLA-4 treatment was assessed. This combination treatment strongly reduced the growth of metastases in both the MC38 and AKTPF models. Treatment resulted in enhanced expansion of activated CD8+ T cells, which had features of tumor-reactive exhausted cells, and there was an increase in their TCR clonal diversity. Combination treatment was also tested in mice bearing PDAC liver metastases (K8484 cells) and induced CRs in all treated mice.
These results suggest that local delivery of IFNα by macrophages in the TME of liver metastases can induce antitumor immunity and improve immunotherapy responses by overcoming resistance mechanisms in the metastatic TME.
Writen by Maartje Wouters, image by Lauren Hitchings.




