It is well known that enzymes catalyze reactions in both directions, and so the ability of proteasomes to not only cleave larger peptides into smaller ones but also splice smaller peptides into larger ones should not be surprising; however, the biological data demonstrating this has been limited. Platteel et al. describe a multi-level approach for identifying immunologically relevant, proteasome-generated spliced epitopes by proteasome-catalyzed peptide splicing (PCPS), and provide insight into the role of PCPS in immune responses.
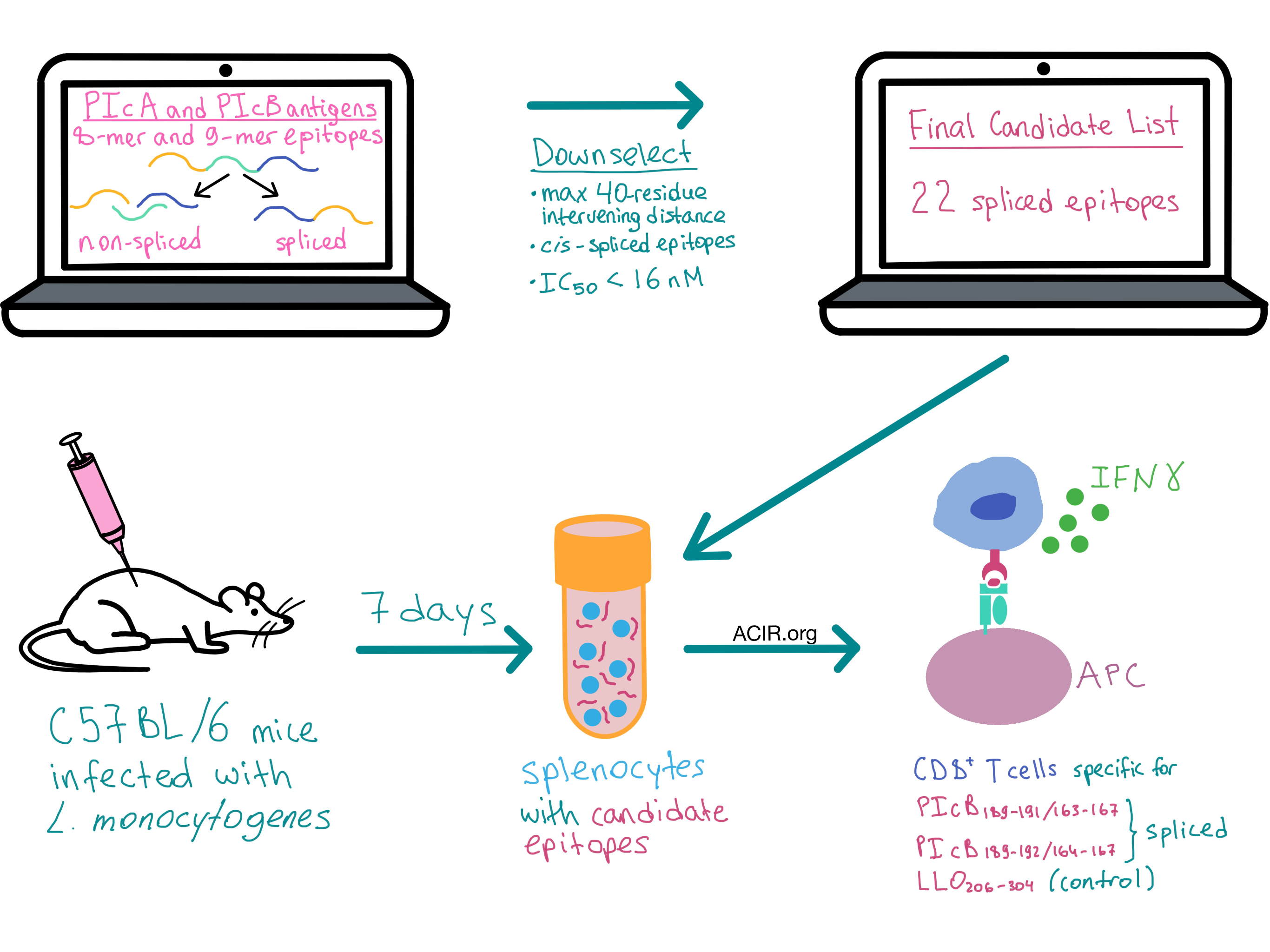
To study how PCPS increases the pathogen-derived peptidome and leads to a broader repertoire of induced CD8+ T cell responses, the researchers utilized C57BL/6 mice infected with L. monocytogenes. During infection, these bacteria secrete listeriolysin O (LLO) and phospholipases PIcA and PIcB in order to access the cytosol of the infected cells. It is known that the LLO296-304 epitope primes CD8+ T cells during infection, however, no PIcA- and PIcB-derived epitopes are known to bind H-2Kb, the C57BL/6 MHC haplotype. Therefore, Platteel et al. chose to use PIcA and PIcB as the model antigens for interrogating peptide splicing in vivo.
The team utilized computational analysis to identify potential spliced epitopes, creating a list of all possible 8-mer and 9-mer non-spliced and spliced epitopes from PIcA and PIcB antigens, and reducing the complexity of spliced epitopes by:
- Specifying a maximum distance of 40 residues between two spliced fragments
- Including only cis-spliced peptides, which are generated from fragments from the same antigen (as opposed to fragments from two different antigens)
The resulting list of non-spliced and spliced peptides was whittled down to 22 peptides with IC50 values below 16 nM (IC50 represents H-2Kb binding affinity, with lower values indicating stronger binding), all of which turned out to be spliced epitopes.
Seven days after L. monocytogenes infection, splenocytes were harvested from mice and incubated with candidate epitopes, and CD8+ T cells were analyzed ex vivo for spliced peptide recognition using intracellular cytokine staining and flow cytometry. Two PIcB spiced epitope candidates (PIcB189-191/163-167 and PIcB189-192/164-167) and the known non-spliced epitope LLO296-304 induced IFN-γ production, indicating an immune response by CD8+ T cells.
Multiple experiments were conducted to confirm the specificity and rule out potential artifacts. Using proteasomes from uninfected (representing the early stages of infection prior to inflammation) and infected (representing changes in the proteasome induced by inflammation) mice in vitro, the researchers confirmed by mass spectrometry that the PIcB-derived spliced epitopes were generated by the proteasome in vitro in both settings. The team also demonstrated that the CD8+ T cells specific for PIcB-derived spliced epitopes did not recognize non-spliced epitopes that shared part of the N or C terminus with the recognized spliced epitopes and that mutation of the anchor residues to non-binding amino acids eliminated reactivity by the cognate T cells. In addition, no closely related (sharing only potential TCR contact residues) non-spliced epitopes could be identified within the entire L. monocytogenes peptidome that could have primed CD8+ T cells specific for PIcB189-191/163-167 and PIcB189-192/164-167. Based on these two results, Platteel et al. concluded that cross-reactivity of these specific T cells against non-spliced epitopes was unlikely to occur.
To validate the use of predicted IC50 as a criterion for selecting potential epitopes, the researchers demonstrated an inverse correlation between the predicted IC50 of the spliced peptides and the frequency of the induced corresponding specific CD8+ T cells. IFN-γ-inducing epitopes PIcB189-191/163-167 and PIcB189-192/164-167 had some of the highest binding affinities of the 22 epitope candidates. Additionally, Platteel et al. observed a significant inverse correlation between the predicted IC50 and the measured half-life of the H-2Kb-peptide complexes, which directly correlated with the relative frequency of epitope-specific IFN-γ+CD8+ T cells. Taken together, these results confirmed the validity of the approach chosen by the authors.
The strategy for identifying the immunologically relevant, proteasome-generated spliced epitopes in vivo described by Platteel et al. provides important biological validation for the role of PCPS in immune recognition of pathogen-derived and self-epitopes and may expand the opportunities to utilize mutation-generated neoantigens for cancer immunotherapy.
by Anna Scherer




