Inspired by the knowledge that the quantity of tumor-infiltrating lymphocytes (TILs) is prognostic for improved survival in breast cancer (BC) patients, particularly those with triple-negative (TNBC) and HER2-overexpressing (HER2+) subtypes, Savas and Virassamy et al., as described in Nature Medicine, set out to uncover whether specific T cell subpopulations affect prognosis in BC patients.
To begin, the team utilized flow cytometry to analyze the TIL content and T cell subsets in tumor samples from 123 patients (84 primary and 45 metastatic samples) with TNBC, HER2+, or hormone receptor-positive (luminal) BC subtypes. They found that TILs constituted a significantly larger percentage of live cells in primary (treatment-naive) than in metastatic tumors across all subtypes, with CD3+ T cells dominating the TIL population. Digging deeper, the researchers found that the most prevalent T cell phenotypes in primary tumors were effector memory (TEM) and terminally differentiated effector memory (TEMRA) subsets.
Given that BC patients generally have limited response to immune checkpoint blockade, with the exception of higher response rates in advanced TNBC (correlating with higher TIL quantity), the researchers analyzed the expression of immune checkpoints in BC TIL subsets. PD-1, but not TIM-3 or LAG-3, was highly expressed on both CD4+ and CD8+ TILs, while CTLA-4 expression was significantly higher on CD4+ than CD8+ TILs. Tregs were infrequent across all BC subtypes, with slightly elevated levels in TNBC.
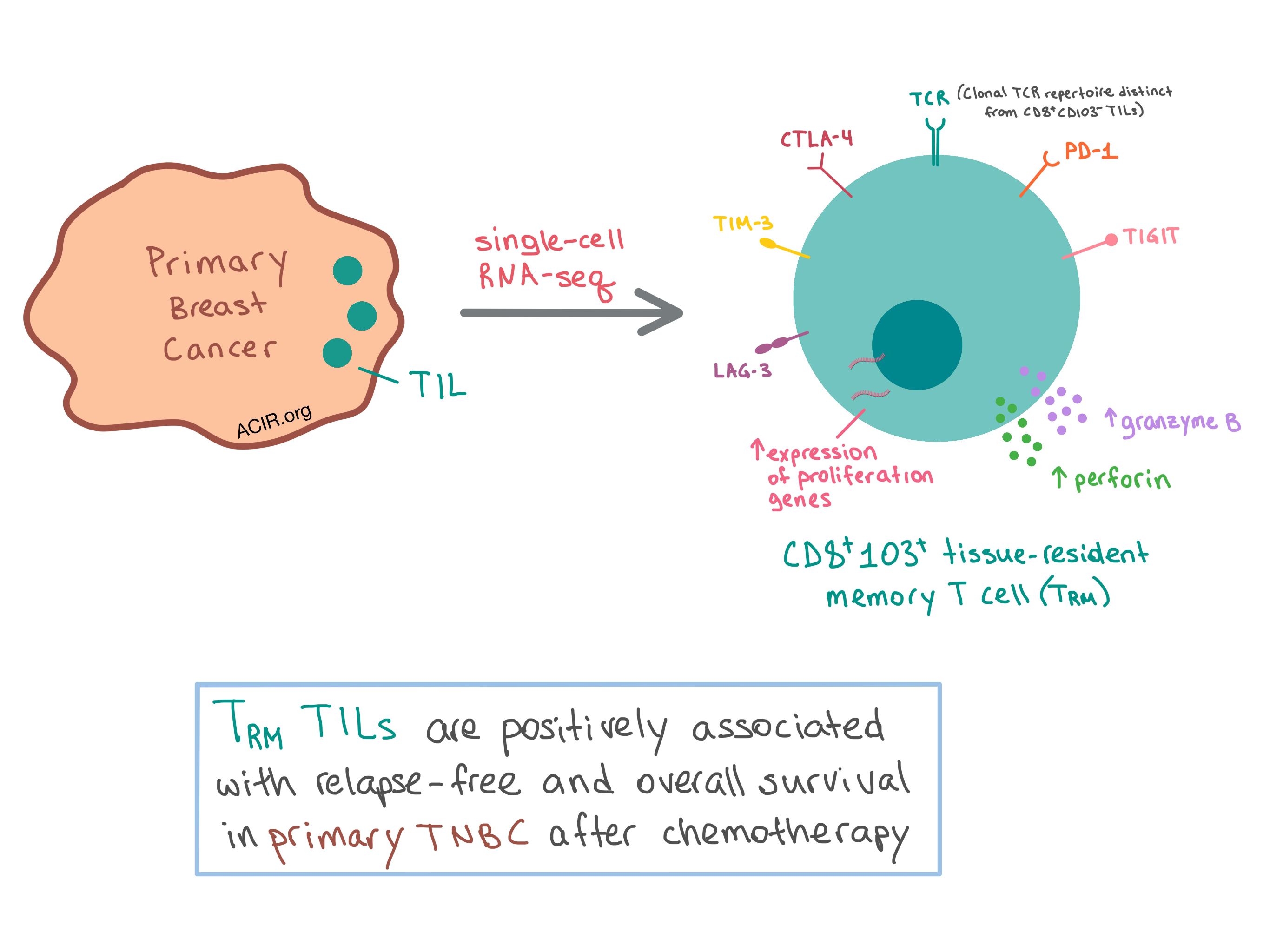
Since TNBC has the highest TIL infiltration among BC subtypes, the researchers chose this subtype to take a closer look at the variation within the T cell infiltrate using a comprehensive single cell mRNA transcriptome analysis. They analyzed 6311 T cells isolated from two human primary TNBC samples and identified ten clusters with unique gene expression profiles, which were similar across the two patients. One cluster that stood out, with 400 differentially expressed genes compared with the other clusters, was the CD8A+CD103+ tissue-resident memory (TRM)-like genotype. These cells had lower expression of tissue egress genes, which is required for retention of the TRM cells within tissue. The TRM cells expressed high levels of immune checkpoints (PD1, TIM3, CTLA4, TIGIT, and LAG3) as well as increased expression of granzyme B and perforin compared with CD8A+CD103- T cells. Another cluster of CD8A+CD103+ T cells was enriched for genes involved in cell proliferation. Overall, the cluster analysis identified a CD8+ TRM-like subset that is cytotoxic and proliferative within the tumor microenvironment.
Expanding their analysis to the entire patient cohort, the team found that a subset of T cells expressed CD103 across all BC subtypes, with significantly more CD8+ T cells than CD4+ T cells expressing this marker. The CD8+CD103+ T cells expressed higher levels of PD-1 and were more frequently double-positive for PD-1 and CTLA-4 than other CD8+ T cell subsets, which was consistent with single-cell analysis. Overall, the data showed that CD8+CD103+ T cells were enriched in BC tumors with a high number of TILs, and bulk RNA-seq of CD8+CD103+ and CD8+CD103- T cells from another three tumor samples confirmed the TRM-like characteristics of the CD8+CD103+ T cells. In vitro co-culturing of TILs with autologous BC cells with or without TCR stimulation revealed that CD8+CD103+ T cells expressed significantly more granzyme B than CD8+CD103- T cells, regardless of TCR stimulation, demonstrating the cytotoxic functionality of this T cell subset.
The team then performed TCRβ sequencing on sorted CD8+CD103+ and CD8+CD103- T cells and revealed distinct TCR repertoires between TRM and TEM subsets in primary tumors, with the former exhibiting higher TCR clonality. These results suggest that the two T cell subsets recognize different antigens. To understand how this came to be, the researchers analyzed normal breast tissue obtained from prophylactic mastectomies and found that the TRM cells make up a significant portion of CD8+ T cells present in the tissue. These observations suggest that the antigen specificity of TRM cells may originate from the microenvironment found in normal, pre-neoplastic, and early neoplastic breast tissue.
Having confirmed the prevalence of the TRM subset in BC TILs, especially primary TNBC, the researchers sought to determine the prognostic value of this subpopulation. Using publicly available gene expression and clinical outcome data from 329 primary TNBC patients, the team found that the TRM gene expression signature was significantly associated with increased relapse-free and overall survival in primary TNBC after chemotherapy treatment.
Overall, Savas and Virassamy et al. demonstrated that a distinct CD8+CD103+ TRM subset of TILs in BC is cytotoxic, proliferative, and clonally enhanced within the tumor microenvironment, and is associated with improved survival in TNBC, pointing to the potential role of the TRM subset as the key player in clinical outcomes that have been observed in BC tumors with a high quantity of TILs.
by Anna Scherer




