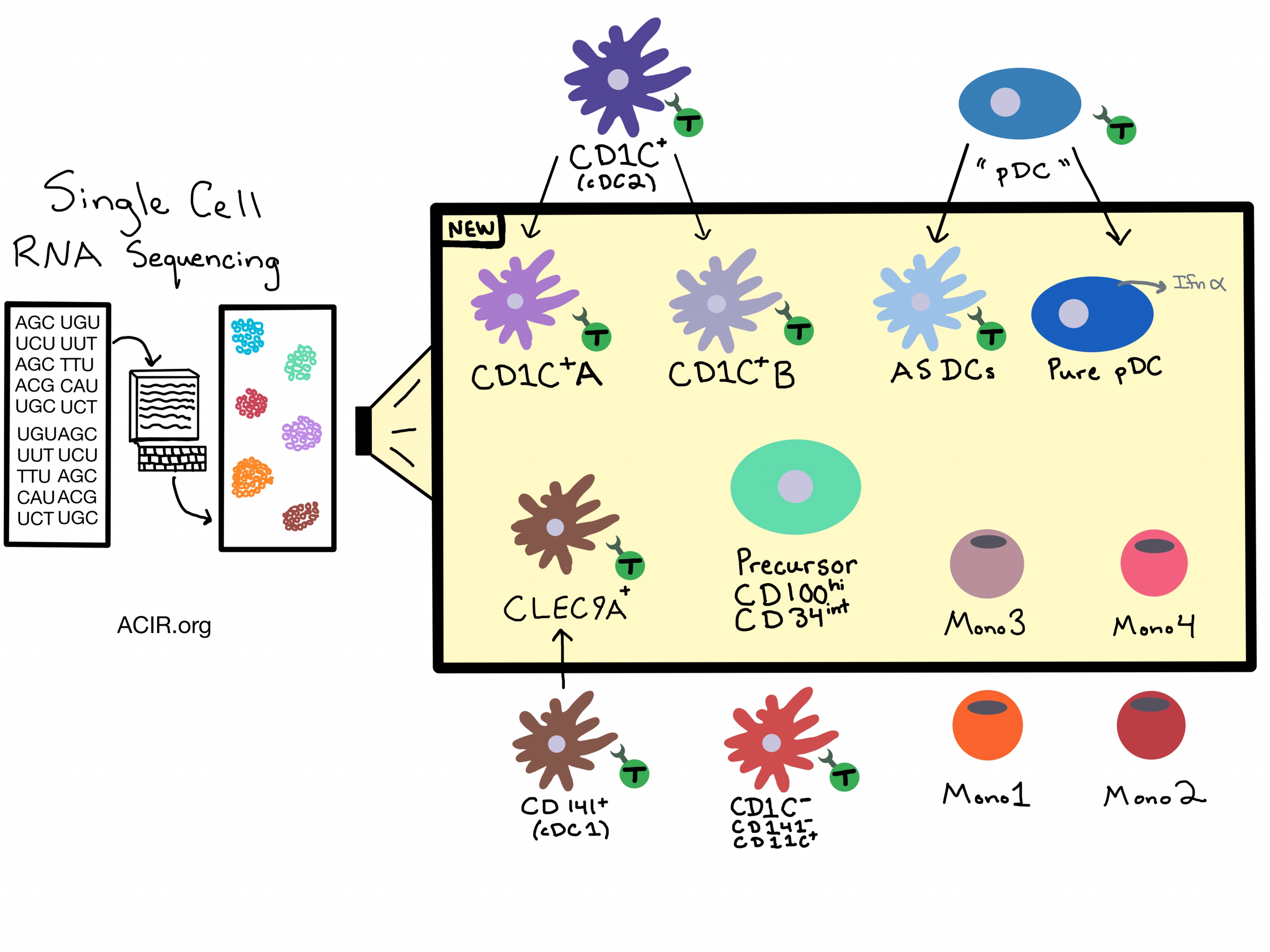
Dendritic cells (DCs) provide the critical nexus between antigenic assault and initiating adaptive immunity. DCs are well known for their antigen presentation capabilities, but when it comes to the classification of DC subtypes, the differences in function, morphology, and expression of a limited number of known surface markers provided useful but limited distinctions. In this study by Villani et al., single-cell RNA-sequencing (scRNA-seq) was used to broadly quantify gene expression patterns and to categorize flow-sorted DCs in an unbiased way; 742 DCs, which were CD14-, LIN- (excluding T cells, B cells, and NK cells), and HLA-DR+ were sampled with an average of 5326 unique genes detected per cell.
Six DC subtypes were identified and validated using the scRNA-seq strategy, broadening the original classification of just four DC subtypes. Previously defined “conventional” cDC1 (CD141+) DCs are now more discriminatively defined as CLEC9A+ DCs and the original cDC2 (CD1C+) class, split into two subpopulations. CD1C+_A is distinguished phenotypically by a higher expression of MHC class II genes, and functionally by secretion of higher levels of immune regulatory cytokines. CD1C+_B is distinguished by a CD14+ monocyte-like, expressed gene set. Both subpopulations potently induce T cell proliferation. Additionally, a new DC subset was identified, expressing AXL, SIGLEC1, and SIGLEC6 antigens, named AS DCs. AS DCs share a similar gene expression signature with plasmacytoid DCs (pDCs), but are functionally and morphologically distinct. AS DCs appear to have been buried in the original pDC population, and the identification of new surface markers allows the “pure” pDCs to be separated from the AS DCs. Functionally, AS DCs potently stimulate T cells and upregulate CD86, while pure pDCs produce IFN-α and do not stimulate T cells. Furthermore, a circulating and dividing precursor of cDCs capable of giving rise to both CLEC9A+ DCs and CD1C+ DCs was identified and defined by the expression of CD100+ CD34int.
Using a similar approach, four clusters of CD14+ LIN- monocytes were distinguished from 339 cells. In addition to the previously defined monocyte subsets, Mono1 (CD14+) and Mono2 (CD16+), two additional monocyte populations, Mono3 and Mono4, were identified. Mono3 expresses a unique combination of genes affecting the cell cycle, differentiation, and trafficking, but its exact functions remain unknown. Mono4 expressed cytotoxic genes in addition to the classical Mono1 gene set.
Accurate classification of DCs with more specific surface markers and improved tools for precise isolation could make DC (or other) vaccines more effective, allowing certain cells with specific functions to be effectively isolated and/or targeted. This improved approach to taxonomy, enabled by single-cell RNA-sequencing, can be extended to other immune and non-immune cell populations.
by Dominique Stavis




