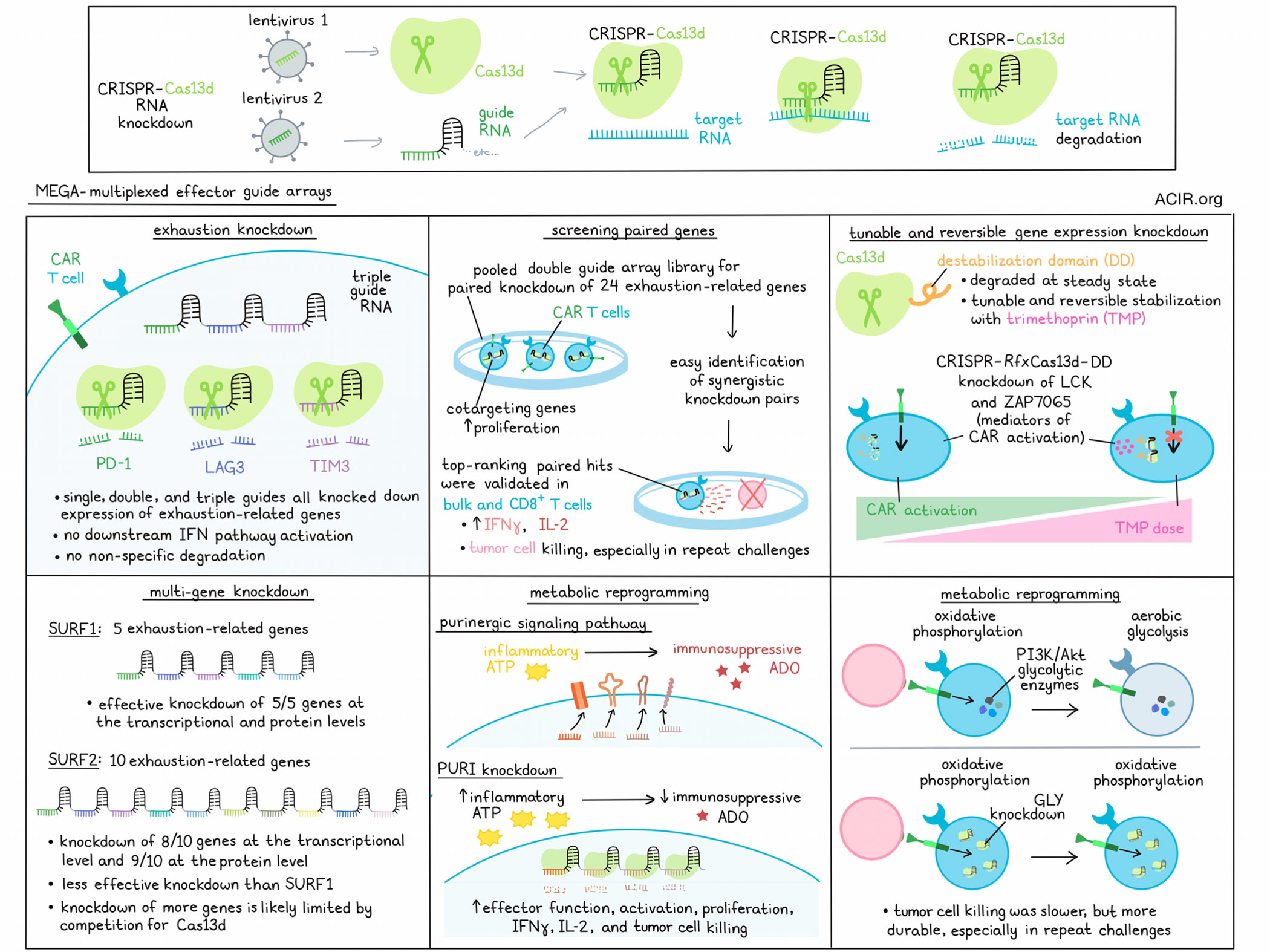
The CRISPR-Cas9 gene editing platform has revolutionized research and medicine by allowing for precise editing of DNA. However, these systems cannot be dynamically regulated and have limitations in their safety and efficacy. To overcome some of these limitations, Tieu et al. developed multiplexed effector guide arrays (MEGA) – a CRISPR-Cas13d platform that targets sequence-specific RNA transcripts for cleavage and degradation, enabling tunable, reversible, and widespread knockdown of selected genes at the transcriptomic level. The team recently demonstrated the versatility and utility of this platform in various settings related to CAR T cell functionality. Their results were published in Cell.
To evaluate the functionality of MEGA, Tieu et al. used it to target LAG3, PD-1, and TIM3, which are typically upregulated in dysfunctional GD2-targeted HA-28ζ CAR T cells. In an optimized manufacturing workflow, primary human T cells were co-transduced with separate RfxCas13d and HA-28ζ CAR lentiviruses. The next day, cells were transduced with CRISPR-associated RNA lentiviruses carrying RNA guides that target specific RNA transcripts in the cell. Single, double, and triple guides targeting LAG3, PD-1, and TIM3 transcripts in various combinations all effectively knocked down expression of those proteins to near baseline levels, with triple combination guides showing various degrees of robustness depending on the spacer configurations between guides. Off-target or downstream biological effects, such as IFN pathway activation and collateral activity (non-specific RNA degradation) were all either minimal or not observed, even when a highly expressed gene such as B2M was selected as a target.
Next, the researchers hypothesized that a pooled double guide array library could be used to easily screen for gene pairings that could be knocked out in tandem, without the Cas9 requirement for barcoding to identify pairs, as compact RfxCas13d guide arrays can be directly sequenced in a single read. Investigating single and double guide RNAs targeting 24 exhaustion-related genes in all 576 pairwise combinations, the researchers found that co-targeting genes generally had a stronger impact on proliferation than targeting single genes. Top-ranking paired hits were functionally validated with guide- and gene-level data, and were experimentally validated in both bulk and CD8+ CAR T cell cultures, which recapitulated screening results and showed evidence of dysregulation within 48 hours after transduction.
Evaluating the impacts of these screen hits on antitumor immunity, the researchers found that top guide RNAs robustly altered IFNγ secretion and moderately improved IL-2 secretion upon tumor stimulation. This improved tumor cell killing across multiple donors and effector:target cell ratios, particularly upon serial stimulation. Notably, while single targeting of FAS or CBLB (both involved in pathways associated with the regulation of T cell activation) had no impact, simultaneous knockdown potentiated strong, durable antitumor responses across multiple tumor rechallenges, demonstrating the utility of evaluating gene knockdowns in tandem.
In the MEGA system, transcript knockdown is dependent on expression of RfxCas13d. In an effort to enable tunable and reversible control of gene knockdown, Tieu et al. fused a destabilization domain (DD) to the C-terminal of RfxCas13d, resulting in rapid degradation of the complex at steady state. However, with the addition of the FDA-approved antibiotic trimethoprim (TMP), the DD is stabilized, allowing for effective Cas13d-DD-mediated cleavage of target RNA. When this system was tested with CD46 as the target gene, the researchers observed completely reversible regulation of gene expression. Further, titration experiments showed that gene expression could be effectively tuned by altering the dosage of TMP.
Taking advantage of the ability to tune RNA expression, the researchers generated a RfxCas13d-DD to target the CAR proximal signaling genes LCK and ZAP7065, enabling tunable regulation of CAR activation. With increasing amounts of TMP in culture, this targeting array reduced CD69 expression, IL-2 secretion, and exhaustion marker upregulation in various CAR T cell models with different costimulatory and scFv domains, demonstrating that MEGA can be used to tune CAR T cell activation in an receptor-agnostic manner.
Investigating just how many genes can be simultaneously knocked down using the MEGA platform, the researchers tested guides targeting 5 (SURF1) and 10 (SURF2) exhaustion-related genes at once. Without optimization, SURF1 knocked down all 5 target genes to baseline levels or lower, and SURF2 knocked down 9 out of 10 genes at the transcriptional level – though not as fully as with SURF1 – and 8 out of 10 at the protein level. These results suggest that the MEGA platform can allow for many genes to be targeted at once, though the researchers hypothesized that guides may begin to compete for Cas13d when higher numbers of genes are targeted.
Next, the researchers developed a guide array targeting expression of four enzymes (PURI) involved in the purinergic signaling pathway, which converts extracellular inflammatory ATP into immunosuppressive adenosine (ADO). Targeting these genes effectively knocked them down, leading to accumulations of ATP, with less generation of AMP and ADO. This improved effector functions in otherwise dysfunctional CAR T cells, increasing their activation, proliferation, production of IFNγ and IL-2, and capacity for tumor killing upon stimulation in vitro.
Similarly, targeting genes for 4 key glycolytic enzymes (GLY) along the PI3K/Akt axis, disrupted the downstream transition from oxidative phosphorylation to aerobic glycolysis in activated T cells, reducing costimulation, Th1 differentiation, and inhibitory markers, and leading to widespread transcriptional reprogramming. The end result was increased oxidative phosphorylation and a fitter, less exhausted phenotype, suggestive of successful metabolic reprogramming. A similar knockdown using CRISPR-Cas9 was much less effective, and showed evidence of potential genotoxicity, suggesting a benefit to using MEGA over traditional CRISPR techniques. Evaluating antitumor efficacy, MEGA-treated GLY cells showed lower activation and secretion of IFNγ and IL-2, with fewer cells expressing IFNγ. While these cells initially cleared tumors at similar or slower rates than control CAR T cells, they outperformed upon repeated stimulations, and significantly enhanced tumor killing over time. Upon transfer into mice bearing Nalm6-GD2 tumors, MEGA-edited cells were more abundant and cleared tumors significantly better than controls.
Overall, these results show that the MEGA platform, which effectively utilizes CRISPR-Cas13d to knock down gene expression at the transcriptomic level, has numerous applications, from screening knockdowns of multiple genes at once, to tuning gene expression and CAR activation, to metabolically reprogramming cells and improving their antitumor abilities. Offering several advantages over CRISPR-Cas9, including tunability, reversibility, and reduced risk of genotoxicity, MEGA will likely serve as a handy tool in both research in medical toolkits.
Write-up and image by Lauren Hitchings




