Few targetable antigens are known for pediatric brain tumors, which have limited immunotherapy development. Based on this pressing need, Raphael et al. analyzed the TCR repertoires of tumor-infiltrating T cells (TILs), and predicted antigens based on these repertoires. Their results were recently published in Science Translational Medicine.
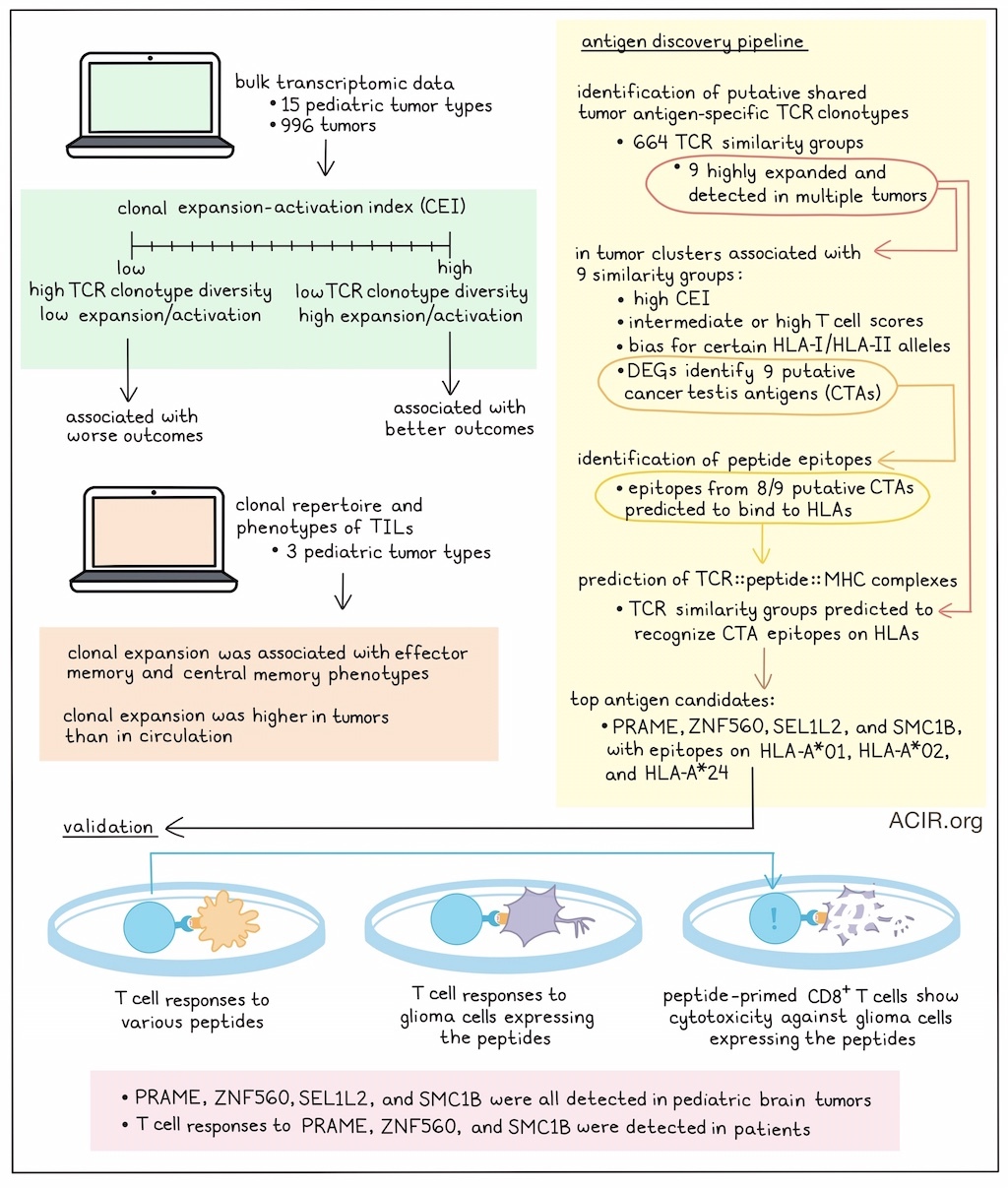
The researchers used bulk transcriptomic data from 996 tumors, representing 15 pediatric tumor types, to perform an in-depth characterization of TCRαβ genes. To estimate clonal diversity, the number of CDR3 clonotypes was normalized to the number of TCR reads in each sample to retrieve the normalized T cell clonal repertoire, representing the proportion of the TCR repertoire consisting of recurring clonotypes. The normalized clonal repertoire correlated with genes related to TCR signaling, T cell proliferation, and T cell activation. Based on this, the researchers named it the clonal expansion-activation index (CEI), representing an estimate of T cell activation and clonality. A high CEI score suggested a relatively high number of TCRs with low clonotype diversity, suggesting tumor-related activation, expansion, and diversity contraction. A low CEI score, on the other hand, suggested the presence of diverse T cell clones with limited expansion and activation.
High CEI was correlated with gene pathways enriched in T cell activation, antigen presentation and processing, and cytokine response. Genes shared across high CEI score-associated pathways included factors previously associated with antitumor immunity in brain tumors, as well as genes related to inhibiting T cell responses.
CEI scores were variable among different tumor types. Patients were stratified into high- and low-risk groups based on CEI scores to determine whether CEI could predict outcomes. High-CEI groups had better prognosis, while low-CEI groups had worse outcomes.
Medulloblastoma has four established molecular subgroups (SHH, WNT, G3, and G4), which are associated with specific genomic signatures, anatomic locations, ages, pathologies, prognoses, and clinical outcomes. Correlating CEI variations within medulloblastoma samples with these molecular subgroups showed that SHH tumors had the lowest CEI and poor outcomes, while WNT had the highest CEI and more favorable outcomes. While there was wide variability in CEI scores in G4 tumors, and G3 tumors had high CEI but poor prognosis, overall stratification based on CEI was more accurate in prognostic risk determination than stratification based on molecular subtypes alone, and G3 and G4 groups could be separated into additional risk groups.
Next, the clonal repertoires and phenotypes of TILs were characterized in three types of pediatric brain tumors using single-cell RNAseq and TCRseq. Various subtypes of antigen-experienced CD8+ TIL, such as Tem, Tcm, exhausted (Tex), and precursor exhausted (Tpex) cells and granulysin CD4+ TIL expressed genes related to antitumor responses. Clonally expanded T cells (defined as having a TCR clonotype shared by ≥3 cells) were associated with Tem- and Tcm-enriched clusters. Higher numbers of clonotypes were detected in circulation, and few of these were shared with the tumor. Higher abundance of expanded TCR clonotypes within the tumor suggested tumor-specific clonal expansion in TILs.
Using an antigen discovery pipeline, the researchers then identified putative shared tumor antigen-specific TCR clonotypes. First, public TCRs and pathogen-specific TCRs were filtered from the TIL TCR clonotypes, and the remaining clonotypes were clustered based on CDR3 sequence homology, length, and shared TRBV genes. This retrieved 664 TCR similarity groups, which may recognize shared or cross-reactive antigens. Nine of these TCR similarity groups (C1 to C9) were detected in multiple tumor types and were highly expanded, suggesting reactivity to a common tumor antigen, were further analyzed.
The researchers found that some HLA-I and HLA-II alleles distributed more prominently within tumor sample clusters related to these TCR similarity groups. Further, most samples within these clusters had high CEI scores and intermediate or high T cell scores, suggesting these similarity groups were detected in immunologically hot tumors.
Next, the transcriptomic landscapes of tumors were assessed to search for potential shared tumor-associated antigens (TAA) recognized by the C1 to C9 TCR similarity groups. A differential gene expression analysis was performed on each cluster, and genes were selected that were not expressed in healthy tissues (excluding testis/reproductive organs) to identify putative cancer/testis antigens (CTAs). Genes with >2-fold change from controls and associated with prognosis were selected, and nine putative antigens were retrieved. An artificial neural network-trained model then identified peptide epitopes from eight of the nine putative antigens that were predicted to bind to HLA-I molecules with high affinity. Subsequently, a deep learning-based model was used to predict the formation of TCR::peptide::MHC complexes (immunologic synapses). This analysis predicted that TCR similarity groups could recognize epitopes presented by HLA molecules beyond those originally associated with the cluster.
The top antigen candidates were PRAME, ZNF560, SEL1L2, and SMC1B, with peptide epitopes for HLA-A*01, HLA-A*02, and HLA-A*24. Raphael et al. assessed their immunogenicity using isolated T cells from healthy PBMCs with corresponding serotypes. Various epitopes induced CD8+ T cell responses, and some of these T cells also produced IFNγ in response to glioma tumor cell lines expressing the peptides. Blocking HLA-I removed these effects. Further, peptide-primed CD8+ T cells induced cytotoxicity against autologous peptide-pulsed target cells and against glioma cells expressing the target genes.
To evaluate whether these antigens might be useful targets in pediatric brain tumors, protein expression of PRAME, ZNF560, SEL1L2, and SMC1B was assessed in 37 tumor samples and 11 nonmalignant brain tissues. These antigens were detected at various degrees in all analyzed tumor types, with only sporadic expression in healthy tissue. To explore whether the predicted epitopes triggered spontaneous antigen-specific responses in TILs and PBMCs of patients with pediatric brain tumors, T cells from 10 patients were stimulated with the predicted peptides. Various patient samples contained SMC1B- or PRAME-specific T cell responses, which were not detected in healthy controls. Two patients had T cells among PBMC and TIL that could recognize ZNF560.
In total, this study found that in pediatric brain tumors, TCR clonality was associated with prognosis, which is an interesting finding regarding potential selection of patients for immunotherapies. Further, the study showed the applicability of a pipeline to predict tumor antigens, which may be further developed for more precise immunotherapies in this patient population with a high unmet need.
Write-up by Maartje Wouters, image by Lauren Hitchings.




