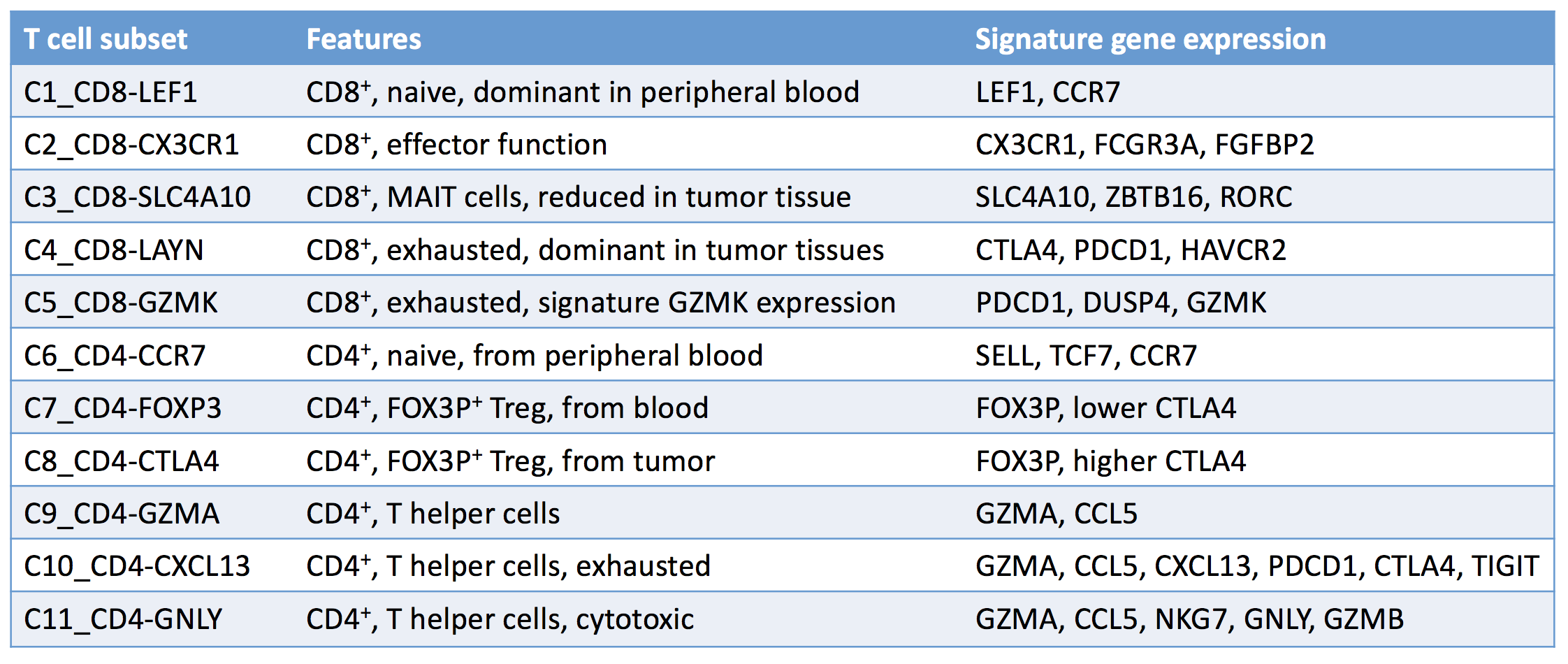
The more data you collect, the more details, and hopefully insights, you’ll uncover; that’s why Zheng et al. performed single-cell RNA sequencing and T cell receptor (TCR) profiling on over 5,000 individual T cells isolated from the tumors, adjacent normal tissue, and peripheral blood of six patients with hepatocellular carcinoma (HCC). From the mass of data collected, the researchers identified 11 unique T cell subsets with distinct patterns of T cell clustering and signature variable genes. Subsets were categorized based on their molecular and functional properties, as well as their developmental trajectories.
Tumor infiltration and TCR clonality
Overall, HCC had poor CD8+ T cell infiltration, making it a good model to study TIL dysregulation. Ten percent of CD8+ T cells found in normal tissue and blood had recurrently identified TCRs, while 30% of T cells from tumor tissues showed such TCRs, indicating clonal expansion, possibly within the tumor. Similar results were observed for T regulatory cells. Within the tumor, clonal CD8+ T cells were also more likely to be exhausted than T cells without TCR clones.
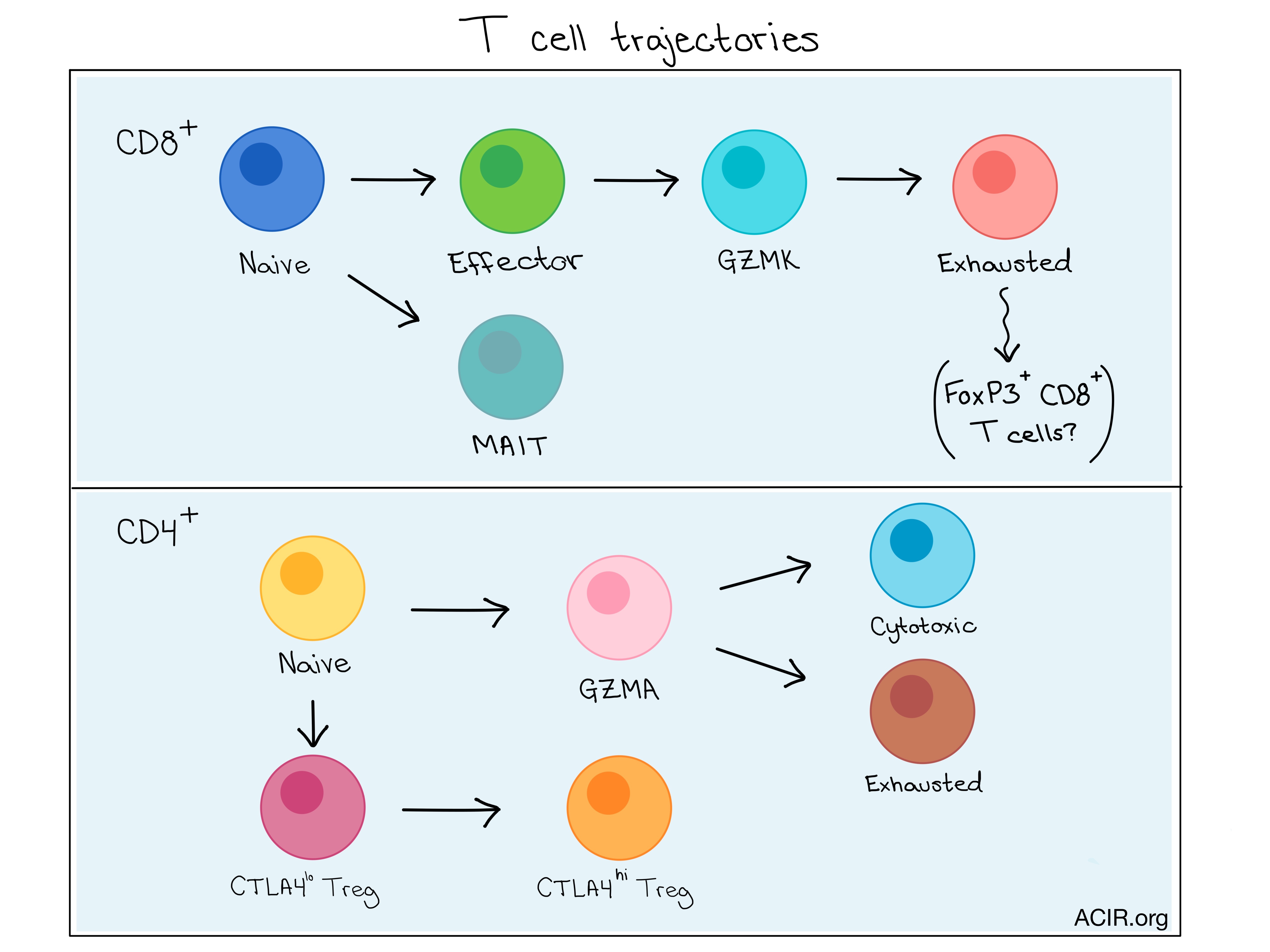
Exhausted CD8+ T cells (C4_CD8-LAYN) and active CTLA4hi T regulatory cells (C8_CD4-CTLA4) were significantly enriched in tumor samples and were potentially clonally expanded, and late stage cancers had higher exhausted CD8+ T cell populations. Meanwhile, percentages of MAIT cells were reduced in tumor samples compared to normal tissue. A small CD8+ cell subpopulation was also found to express the FOXP3 T regulatory marker, suggesting a possible developmental relationship between Tregs and exhausted CD8+ T cells.
Exhaustion
By comparing exhausted and non-exhausted cytotoxic CD8+ T cells, Zheng et al. identified 82 exhaustion-specific genes, including well-described markers like CTLA4, and PDCD1, along with less-described markers including MYO7A, WARS, and CXCL13, and novel exhaustion markers including LAYN, PHLDA4, and SNAP47. Interestingly, twenty-two of the exhaustion genes identified in cytotoxic T cells were also highly expressed on tumor-infiltrating Tregs.
Zheng et al. took a closer look at the LAYN gene and found that LAYN was upregulated both in activated Tregs, especially those with a more repressive phenotype, and exhausted CD8+ T cells isolated from the HCC tumor, and that high LAYN expression was associated with reduced disease-free survival. Through in vitro studies, they also showed that LAYN was only expressed following CD3 and CD28 activation of CD8+ T cells, and once expressed, it acted to repress CD8+ T cell functions. When CD8+ T cells were induced to overexpress LAYN, they produced significantly less IFN-γ. Interestingly, LAYN was expressed on LAG-3-, but not LAG3+, CD8+ T cells – a mutual exclusivity that could indicate distinct T cell subsets.
Differentiation trajectories
Combining the information from complete transcriptome and TCR analyses allowed the researchers to gain insights into the functional states of, and relationship between T cell subsets. Using algorithms to map the differentiation trajectories based on changes in expression over “pseudotime”, CD8+ T cells were shown to move sequentially from a naive to an effector state, then to an exhausted state expressing GZMK, and finally to a terminally exhausted state. This suggests that GZMK expression represents an intermediate state en route to exhaustion.
Analysis of the CD4+ T cell trajectory revealed that naive CD4+ and CD4+ T helper cells did not separate over pseudotime until T helper cells branched into either an exhausted or cytotoxic state, indicating functional divergence of these derivative subsets. The two end states were more closely related to the intermediate T helper state than to one another, as revealed by the number of shared TCRs between subsets.
In order to contribute further to the base of scientific knowledge and help other scientists out, Zheng et al. developed an interactive web-based tool for analyzing, visualizing, and downloading their single-cell data for single or multiple user-input genes. The tool can be found at: http://hcc.cancer.pku.cn.
Overall, exploring the function and dysfunction of T cells in the tumor microenvironment is critical to understanding mechanisms of action and identifying useful biomarkers and targets for cancer immunotherapies.
by Lauren Hitchings




