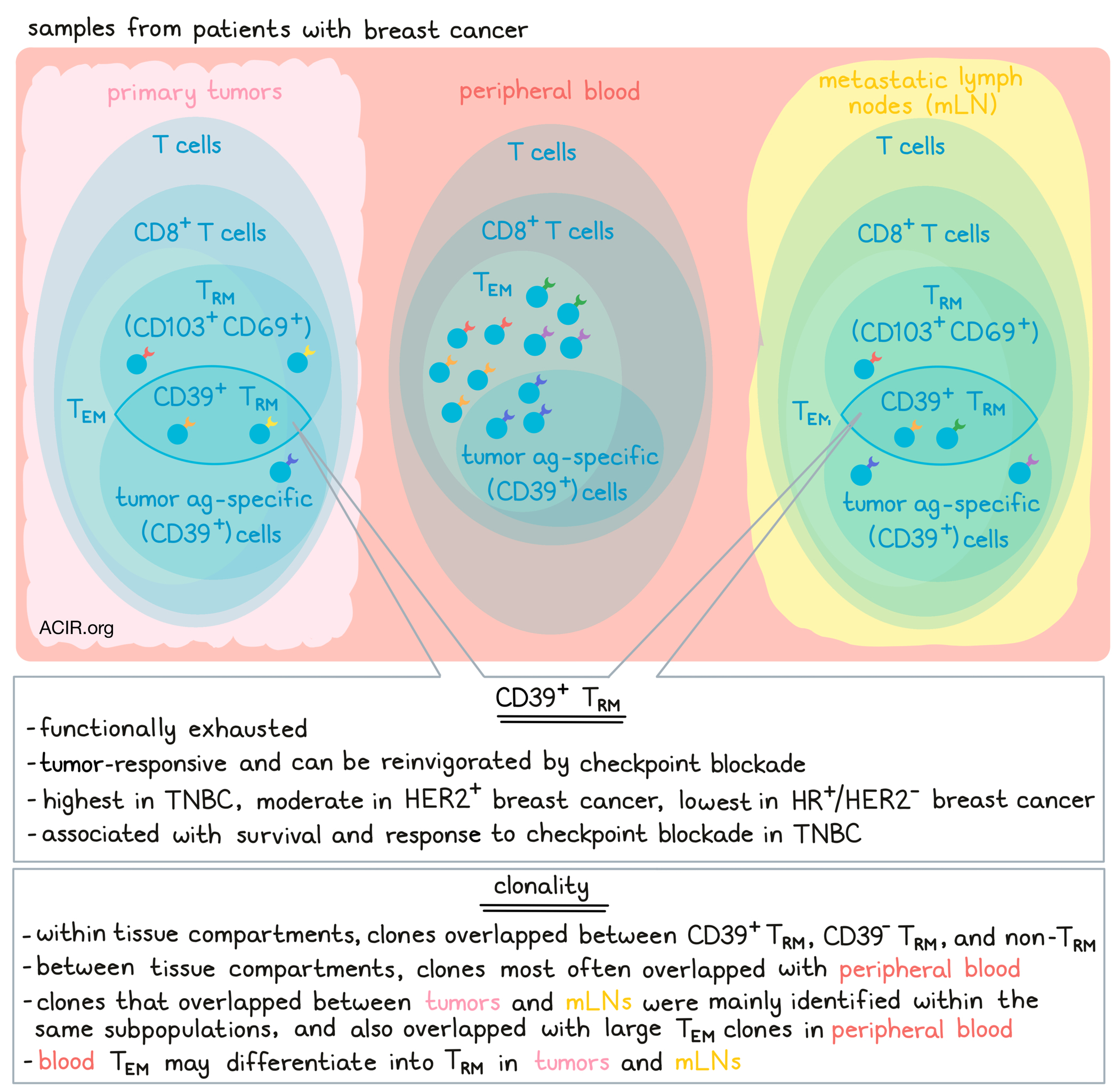
Investigating the importance CD39+CD8+ T cells in prognosis and in the success of immune checkpoint blockades for breast cancer, Lee et al. examined the heterogeneity of CD8+ T cells obtained from different compartments, including primary tumors, metastatic lymph nodes (mLNs), and peripheral blood, and characterized cellular phenotypes and functionalities of T cells using data from multicenter cohorts of patients with breast cancer who underwent curative aimed-surgery. Their results were published in Science Immunology.
To begin, the team looked at the composition of immune cells in peripheral blood and tumor tissues from 131 patients with early breast cancer. Among CD45+ cells, T cells were the most abundant cell type in tumors. Among T cells, the percentage of CD8+ cells was higher in tumors than in blood, as was the percentage of CD8+ T cells expressing CD69 and CD103 – markers of tissue-resident memory T cells (TRM). CD103 was later found to be exclusive to CD69+CD8+ T cells, and CD103 was subsequently used to define TRM.
Investigating the antigen specificity of T cells within tumors, the researchers first identified virus-specific bystander T cells. While bystander T cells recognizing influenza A virus (IAV)- were enriched in tumors over peripheral blood, those recognizing human cytomegalovirus (HCMV) were not, possibly because a higher percentage of IAV-specific T cells were CD69+CD103+ TRM. Next, the team identified T cells recognizing NY-ESO-1, which is frequently expressed in triple-negative breast cancer (TNBC). These cells could be found in the tumors and blood of patients with TNBC. Within the tumor, but not the peripheral blood, NY-ESO-1-specific CD8+ T cells were enriched for TRM relative to all CD8+ TILs.
Next, the researchers evaluated expression of CD39, which is typically expressed in tumor antigen-specific T cells. Here, CD39 expression was increased among TRM vs non-TRM. Sorting CD8+ TILs into subpopulations based on tissue residency and tumor antigen specificity, the researchers defined three subpopulations: CD39+ TRM, CD39- TRM, and non-TRM. Gene clustering analysis, Gene Ontology (GO) biological pathway analysis, and gene set enrichment analysis together revealed features of functional exhaustion in CD39+ TRM, including upregulated expression of genes related to exhaustion, immune checkpoints, effector functions, and activation, and downregulation of genes associated with naive or central memory CD8+ T cell phenotypes. Genes upregulated in CD39+ TRM were also more characteristic of previously identified TRM signatures, whereas CD39- TRM were more effector memory T cell-like. Upon activation, CD39+ TRM produced less IFNγ, TNFα , and IL-2, and were less polyfunctional compared to other subpopulations. Similar results were observed in cells from mLNs.
Turning their attention towards clonality, Lee et al. performed TCR analysis and compared CD8+ T cell populations between primary tumors, mLNs, and peripheral blood. Defining cells as naive, (TN), central memory (TCM), effector memory (TEM), or terminally differentiated effector memory (TEMRA), the team found that TEM were predominant in tumors and mLNs. Further, many TRM were detected within the TEM population. Within each compartment, some clonotypes overlapped between CD39+ TRM, CD39- TRM. and non-TRM, with minimal effects of contaminating minor clonotypes on TCR clonotype analysis. Between compartments, anywhere from 9-62% of clonotypes found in tumors or mLNs were also detected in other compartments (intercompartmental), with over 80% of intercompartmentally overlapping clonotypes found in peripheral blood. Clones that overlapped between tumors and mLNs were mainly identified within the same subpopulations (CD39+ TRM, CD39- TRM, or non-TRM), and mainly overlapped with TEM as highly expanded (large) clones in the blood. Further, blood TEM clonotypes were distributed among various CD8+ T cell subpopulations, including CD39+ and CD39- TRM, suggesting that blood TEM may differentiate into TRM in tumors and mLNs.
Using a coculture system, Lee et al. confirmed that CD39+ T cells recognized tumor antigens and were reactive against tumor cells. Tumor-reactive (upregulated 4-1BB) TRM were then sorted and TCR sequenced. Mapping these cells back to the patient, the researchers found that tumor-reactive 4-1BB+CD39+ TRM clonotypes with inter-subpopulation clonal overlap in culture were readily detected in blood TEM cells as large clones and broadly distributed among various subpopulations in tumors and mLNs compared with those without inter-subpopulation clonal overlap, confirming systemic clonal overlap.
Lee et al. then examined CD39+ TRM enrichment in early breast cancers with different molecular subtypes: HR+/HER2-, HER2+, and TNBC. While the portion of CD8+ among CD3+ cells and CD103+ among CD8+ cells was consistent across molecular subtypes, the portion of CD39+ among CD8+ cells varied, with the highest enrichment of CD39+ cells in TNBC and the lowest in HR+/HER2- breast cancer. Applying a gene signature for CD39+ TRM to data from The Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) and The Cancer Genome Atlas (TCGA) also showed that that CD39+ TRM were highest in TNBC and lowest in HR+/HER2- breast cancer, including in a metastatic tissue cohort. They also found that while the CD39+ TRM score did not correlate with survival in HR+/HER2- or HER2+ breast cancer cohorts, it did strongly correlate with better cancer-specific survival in TNBC, indicating that CD39+ TRM may directly contribute to antitumor immune responses.
To determine whether CD39+ TRM could be reinvigorated by checkpoint blockade, the researchers performed ex vivo functional analysis assays using CD8+ TILs from a patient with TNBC and NY-ESO-1-specific CD8+ T cells. Here, stimulation with NY-ESO-1 peptides induced proliferation and cytokine release in CD39+ TRM, but not CD39 TRM or non-TRM, and these effects were enhanced with anti-PD-1 and with the addition of anti-CTLA-4. Similar results were observed when CD8+ TILs from another patient with TNBC were cultured with autologous EPCAM+ tumor cells. While restoration of functionality with checkpoint blockade was limited in samples from HR+/HER2- breast cancer, a range of results were observed in samples from patients with HER2+ breast cancer or TNBC, and restoration correlated with enrichment of CD39+ TRM, suggesting that these cells are likely a responding subpopulation that can be functionally restored.
Finally, comparing single-cell RNAseq data from a clinical trial of neoadjuvant anti-PD-1 in patients with breast cancer to their bulk RNAseq data, the researchers found that upregulated genes in expanding CD8+ TILs were also enriched in CD39+ TRM over CD39- TRM. Similarly, applying their CD39+ TRM signature against public scRNAseq dataset from a trial of anti-PD-L1 in patients with advanced TNBC, they found upregulation of CD39+ TRM signature genes in CXCL13+CD8+ T cells, which were associated with response to PD-L1 blockade.
Together, these results show that enrichment of CD39+ TRM, especially those that clonally overlap across compartments, may contribute to survival and response to checkpoint blockade in some patients with breast cancer. These findings may be useful in evaluating therapeutic options for such patients.
Write-up and image by Lauren Hitchings




