Chronic diseases, such as HIV and cancer, are typically associated with an “exhausted” class of T cells characterized by poor function in disease control. Based on the hypothesis that a better understanding of the phenotypic variation among exhausted T cells may more effectively guide immunotherapy approaches, Bengsch et al. set out to systematically categorize exhausted T cell types in different disease settings using a systems immunology approach. The results, published in Immunity, revealed numerous epigenetically defined exhausted T cell subsets with defining features of both disease and anatomical location.
Bengsch et al. began by analyzing gene expression changes in the classically studied chronic lymphocytic choriomeningitis virus (LCMV)-specific exhausted T cells (Tex) compared with naive, effector, and memory T cells after 30 days of infection in order to identify a core exhausted cell gene expression signature. The set of genes with increased or decreased expression, which included CTLA-4, PDCD1, CD38, and ENTPD1 (encoding CD39), was further validated by gene set enrichment analysis. This gene signature was also strongly enriched in tumor-infiltrating lymphocytes (TILs) from melanoma patients and in HIV-specific T cells from HIV progressor patients, suggesting that it may be applicable across disease states.
As epigenetic patterns may be more accurate in describing cell identity than gene expression, Bengsch et al. explored the epigenetic changes in Tex cells in LCMV public data sets using ATAC-seq, which examines the accessibility of open chromatin regions (OCRs). Some of the genes with increased expression and OCR accessibility in Tex cells included inhibitory receptors (Pdcd1, Tigit, Ctla4), ectoenzymes involved in metabolic regulation (Cd38, Entpd1), chemokines and cytokines (Xcl1), and transcription factors (Eomes, Ikzf2, Tox). Genes with decreased expression and OCR accessibility included Ccr7, Il7r, Nt5e, Tcf7, and Lef1. The researchers concluded that this set of genes may serve as a biomarker for exhaustion.
Next, the team sought to apply the population-based epigenomic exhaustion signature to profile Tex cells on a single-cell basis. To do this, they created a mass cytometry panel of 16+ exhaustion-related gene products and other T cell markers of lineage and differentiation state for analysis by cytometry by time of flight (CyTOF). This subset selection was validated via gene set variation analysis of a published CD8+ T cell single-cell transcriptomic dataset from human melanoma tumor-infiltrating lymphocytes (TILs); the validation confirmed that this subset of genes was representative of key features of exhaustion.
After validation, Bengsch et al. applied the CyTOF panel to peripheral blood mononuclear cells (PBMCs) from healthy control subjects, HIV patients on and off antiretroviral therapy, and samples from patients with lung cancer (including PBMCs, macroscopically uninvolved lung tissue, and TILs). They found that PD-1+CD8+ T cells expressed more exhaustion markers than any other CD8+ T cell phenotypes examined. In HIV, molecules predicted to be decreased in Tex correlated with health and mild disease, while molecules predicted to be increased in Tex correlated with advanced disease.
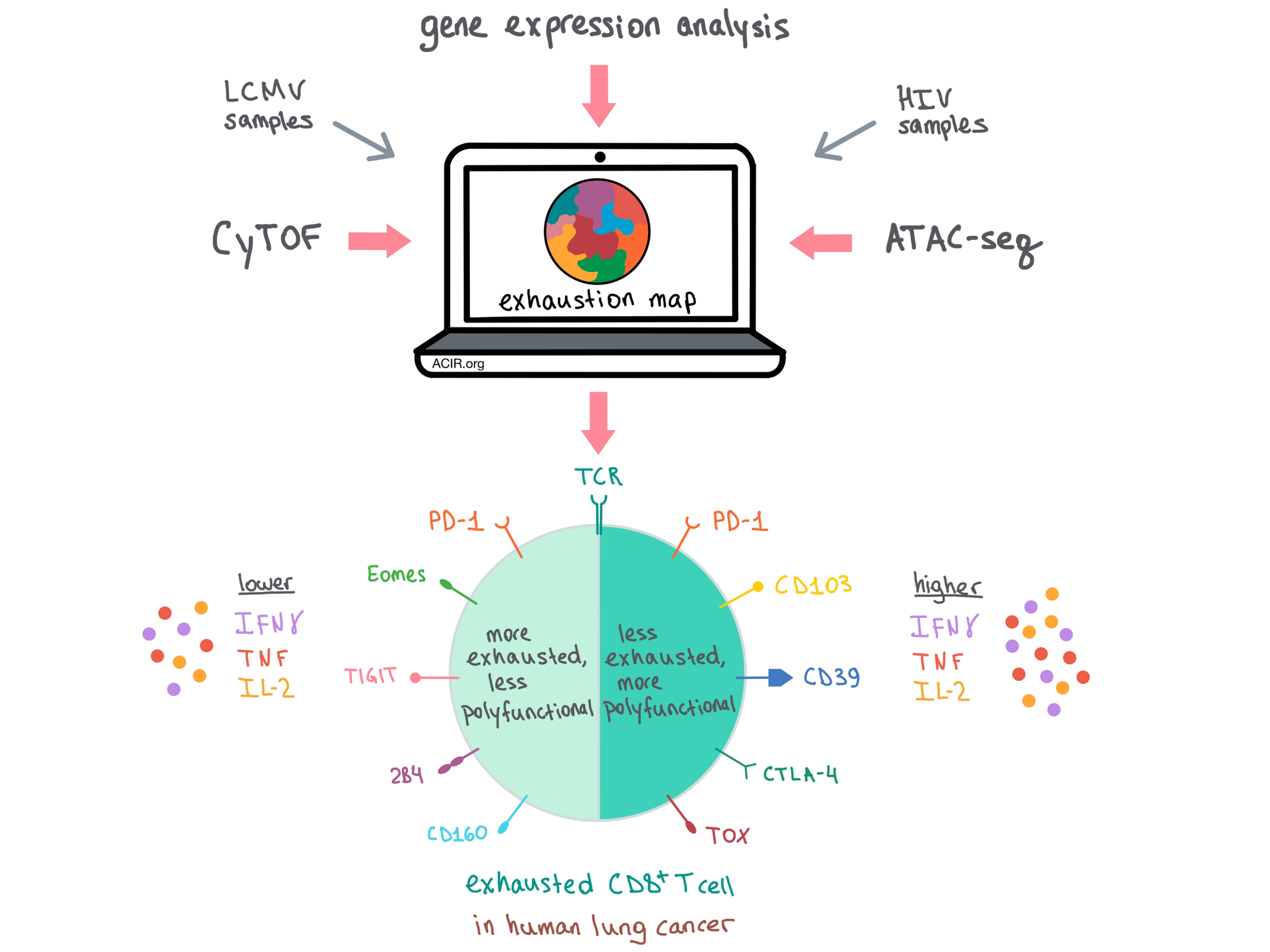
The team then performed a high-dimensional analysis on CyTOF-analyzed non-naive CD8+ T cells to create an “exhaustion map��� and showed that the pattern of marker co-expression differed among multiple Tex cell phenotypes and across patients and diseases. Using a previously developed method, phenograph, the researchers then dug deeper into the data and developed a functional exhaustion score (FES) that increased with hallmarks of exhaustion (e.g., loss of IFNγ, TNF, and IL-2 production) and decreased with the presence of effector or memory functionality to distinctly score each population.
Distinguishing Tex cells from effector T cells using individual markers has been a challenge historically, as some markers of activation also ultimately indicate exhaustion. Using high-dimensional analysis, Bengsch et al. were able to distinguish the two T cell populations by taking into account multiple markers and FES at the same time. Effector T cells had higher expression of CD39, LAG-3, Helios, and CTLA-4, while Tex cells upregulated Eomes, TOX, 2B4, and TIGIT. TCF1, a critical transcription factor in naive and memory cells, was primarily observed in non-exhausted cells, although it was also observed in two exhausted subpopulations.
To find out whether the key features of exhaustion exist across different diseases and tissues, the team analyzed TILs from patients with newly diagnosed lung cancer, and found conserved Tex cell biology across HIV and lung cancer, along with some disease-specific features. Drilling down further into the functionality of these populations in lung cancer, TILs with low IFNγ production co-expressed PD-1 as well as markers of more severe exhaustion, including Eomes, TIGIT, 2B4, and CD160. Meanwhile, the more functional TILs with higher IFNγ production also expressed PD-1, as well as CD103, CD39, CTLA-4, and TOX, but lacked the more severe exhaustion features.
Overall, Bengsch et al. developed a comprehensive systems immunology approach to explore the variation in Tex cells across human diseases, which could provide insights into disease progression and help identify appropriate therapy for patients with specific expression of checkpoint blockade targets.
by Anna Scherer




