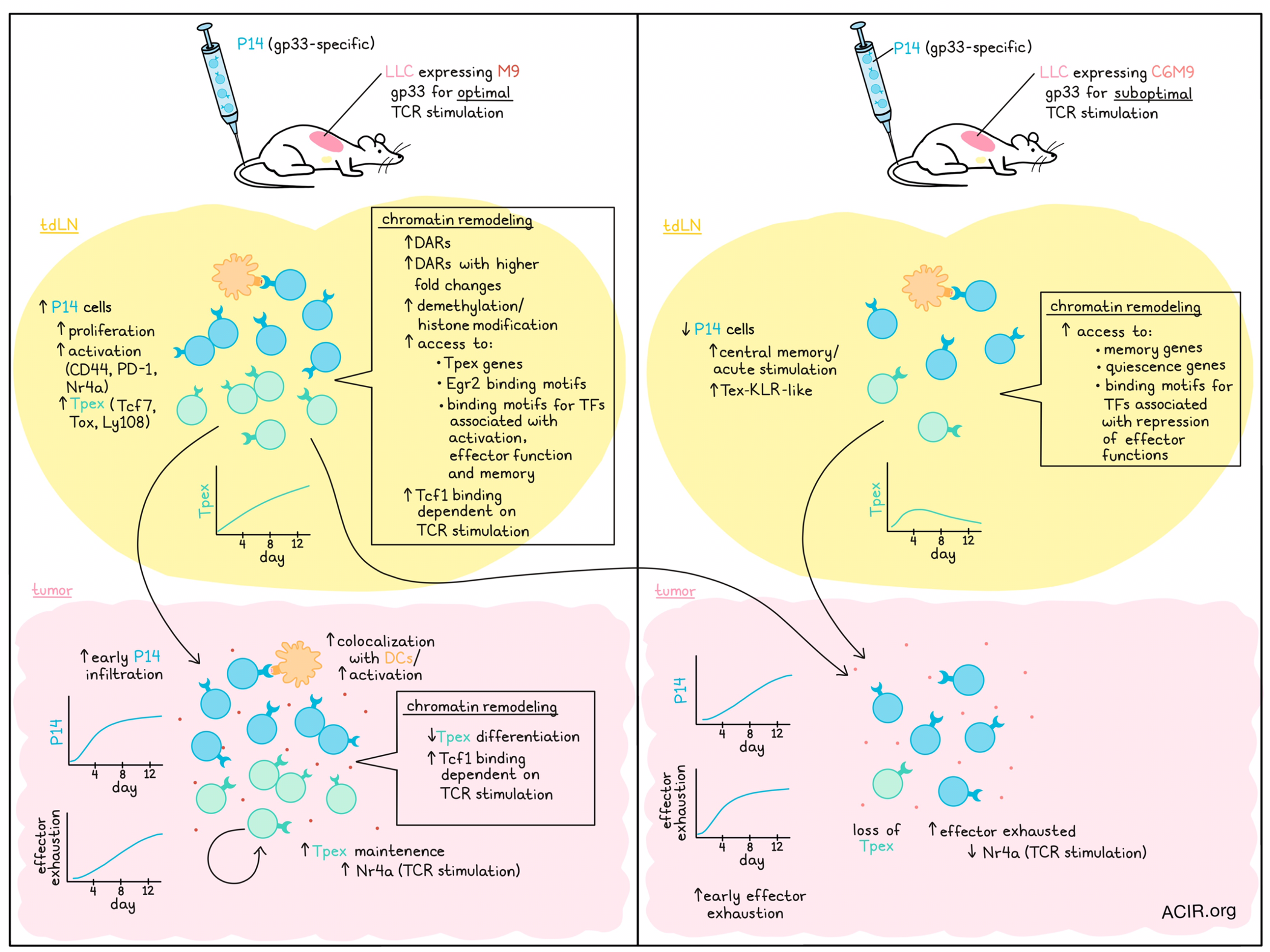
Progenitor exhausted T cells (Tpex), marked by high expression of Tcf1, Tox, and Ly108, help to replenish and maintain effector T cells in tumors, allowing for strong and sustained antitumor functions, including in the context of immunotherapy responses. However, exactly how these cells acquire and maintain this functional phenotype, and whether TCR engagement plays a role is not fully understood. To better understand Tpex cells, Lan et al. developed Lewis lung carcinoma (LLC) models expressing versions of the gp33 antigen that either optimally (M9) or suboptimally (C6M9) engaged the TCRs of gp33-specific T cells (P14). The results of their investigation were recently published in Nature Immunology.
To begin, Lan et al. implanted tumors expressing either M9 or C6M9 into wild-type and Rag-/- mice, followed by adoptive transfer of P14 cells 6 days later. In wild-type mice without P14 cells, the different tumors showed comparable growth, but in Rag-/- mice adoptively transferred with P14 cells, the M9-expressing tumors were better controlled. In wild-type mice with M9 tumors treated with P14 cells, P14 cells in tumor-draining lymph nodes (tdLNs) were more abundant, more proliferative, and expressed higher levels of activation markers CD44, PD-1, and Nr4a compared to in mice with C6M9 tumors. While Tpex cells were induced in both models, they became more enriched through day 12 in the M9 model, while they peaked at day 4 and then reduced in the C6M9 model, suggesting that robust TCR signaling preserved the Tpex phenotype in tdLNs.
Looking at the transcriptomic level, the researchers performed scRNAseq on P14s from the tdLN of tumor-bearing taken 8 days after transfer. Cells from M9-tdLN largely fell into clusters where Tpex gene signatures were highly enriched, while cells from C6M9-tdLN were slightly more proliferative, and fell more into central memory or acutely stimulated clusters, including a cluster that resembled Tex-KLR cells, found in chronic viral infections. Terminally differentiated cells were largely absent in tdLNs from either model.
Next looking at tumors, Lan et al. found that P14 cells more readily infiltrated M9-expressing tumors compared to C6M9-expressing tumors, and were more abundant in early tumors, though the discrepancy decreased with time. Cells in both tumors adopted exhaustion signatures; however, a higher proportion of Tpex cells was maintained in M9-expressing tumors, while a higher proportion of effector-like exhausted cells was established in C6M9-expressing tumors earlier on. Tpex showed higher Nr4a expression, indicative of stronger TCR signaling than exhausted T cells, suggesting that strong TCR signaling may help to maintain the Tpex phenotype in tumors.
Blockade of lymphocyte egress from tdLNs using FTY720 showed that Tpex frequency was reduced, but not lost in M9 tumors, with minimal effects in C6M9 tumors, suggesting that Tpex are both replenished from tdLNs and maintained in the TME. Supporting this, the researchers observed that when P14 cells that had been optimally primed in M9-tDLNs (about 90% Tpex) were adoptively transferred into mice bearing C6M9 tumors, they mostly lost the Tpex phenotype, instead transitioning into more terminal differentiation states with higher effector functions.
Using immunofluorescence imaging to interrogate the tumor immune landscape further, Lan et al. noted that a higher proportion of P14s in M9-expressing tumors colocalized with DCs, and became more activated when they were located adjacent to DC and B cell zones. This effect was not identified in C6M9-expressing tumors, suggesting that sustained optimal TCR engagement may be mediated through enhanced antigen-presentation by APCs.
Digging deeper into how TCR stimulation supports Tpex development and maintenance within cells, the researchers performed ATACseq to evaluate chromatin remodeling. At day 8, the epigenetic landscapes between antigen-experienced P14s from tdLNs in M9 and C6M9 models were distinct, as were the landscapes between antigen-experienced P14s from tdLNs and tumors from the same mice, suggesting that epigenetic programming is affected both by the quality of TCR engagement during priming, and by the tumor environment later on.
Looking at reprogramming related to priming in tdLNs, the researchers found that P14s in M9-tdLNs acquired more differentially expressed regions (DARs) and more DARs with higher fold changes. In Gene ontology enrichment analysis using the top 500 M9-tdLN-DARs, Tpex-associated genes were more accessible, while genes consistent with memory and quiescence were more accessible in P14s from C6M9-tdLNs. Further, M9-tdLN-DARs were enriched for the transcription factor binding motifs of Egr2 (Tex maintenance), and transcription factors that control activation-, effector-, and memory- associated genes. C6M9-tdLN-DARs, on the other hand, were enriched for motifs of transcription factors that repress CD8+ effector T cell functions.
Investigating whether TCR stimulation-induced chromatin remodeling was due to or accompanied by DNA methylation, the researchers performed whole-genome bisulfite sequencing (WGBS). Mirroring the chromatin remodeling data, methylation patterns were distinct between models and tissues, with more differentiated P14 cells from tumors undergoing progressive methylation reprogramming over time. In M9-tdLN, optimally primed P14 cells gained a higher number of demethylated regions (compared to P14s in C6M9-tdLN); were enriched for pathways involved in T cell activation, differentiation, and phospholipid transportation; and were hypomethylated at Tpex genes, corresponding with enhanced chromatin accessibility. Differentially methylated regions were also enriched for sites for AP1 family members and Egr1/Egr2 binding. Despite all this, the majority of differentially accessible regions (DARs; 87.6%) associated with differential TCR signal strength could not be linked to a significant change in DNA methylation, suggesting that histone modification is the predominant mechanism behind the chromatin remodeling.
While the differences in chromatin accessibility in P14 cells in tdLNs between M9 and C6M9 models were more pronounced, the differences in P14s infiltrating the tumors themselves were less distinct. Analysis of DARs suggested that Tpex cells from M9-expressing tumors were less differentiated, and showed little overlap with DARs from tdLNs. Enriched TF motifs were also distinct between models and tissues. Interestingly, Tcf1 was enriched in both the tumors and tdLNs of mice bearing M9-expressing tumors, and the openness of its binding sites were largely dependent upon TCR stimulation. Using CRISPR–Cas9 to disrupt the Tcf1 motifs in a region of Slamf6 (encoding Ly108) induced by TCR-stimulation, Ly108 expression and effector T cell differentiation upon adoptive transfer and tumor infiltration were abrogated, validating the functional relevance of Tpex-specific Tcf1 binding in maintaining tumor-reactive Tpex.
Overall, these results show that optimal TCR engagement plays a role in epigenetic imprinting that supports both Tpex formation during priming and Tpex maintenance and self-renewal in tumors. The effect in tumors was associated with proximity to DCs. Further, suboptimal TCR engagement could inhibit optimally primed Tpex cells, suggesting that maintaining optimal TCR engagement in tumors may be critical to antitumor efficacy.
Write-up and image by Lauren Hitchings




